Prediction of MHC Binding Peptides and Antigenic Peptides from Ascaris lumbricoides
Sonu Mishra and Virendra S Gomase
Sonu Mishra* and Virendra S Gomase
Department of Biotechnology, Mewar University, Chittorgarh, India
- *Corresponding Author:
- Sonu Mishra
Department of Biotechnology
Mewar University
Chittorgarh
India
Tel: 9560808369
E-mail: sonumishra1014@gmail.com
Received date: March 15, 2016; Accepted date: May 18, 2016; Published date: May 25, 2016
Citation: Mishra S, et al. Prediction of MHC Binding Peptides and Antigenic Peptides from Ascaris lumbricoides. J Autoimmun Disod. 2016, 2:2. DOI:10.21767/2471-8513.100018
Copyright: © 2016 Mishra S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The parasitic disease Acariasis is the major concern because of its morbidity and mortality issue. In this investigation, we predicted the binding peptides of the MHC class I and MHC class II by Position Specific Scoring Matrices (PSSM) and Support Vector Machine (SVM) algorithms. We predicted the binding affinity of Cytochrome c oxidase subunit 2 (mitochondrion) having a 232 amino acids long residue sequence, which shows 224 nonamers. We predicted the peptide binding affinity to MHC I moles are as 8mer_H2_Db(The binding thresholds: 33.04,optimal score:52.494), 9mer_H2_Db(Optimal Score: 50.232, Binding Threshold: 17.96),10mer_H2_Db (The Optimal Score: 58.858, Binding Threshold: 41.32), 11mer_H2_Db(Optimal Score: 79.495, Binding Threshold: 56.96) and MHC- II molecules are as I_Ab.p, I_Ad.p, I_Ag7. We also study integrates prediction of peptide MHC class I binding; proteasomal C terminal cleavage and TAP transport efficiency by using sequence and properties of the amino acids. We also found the binding of peptides to different alleles by using Position Specific Scoring Matrix. PSSM based server will predict the peptide binders of Cytochrome c oxidase subunit 2 from Ascaris lumbricoides sequence to, which are found antigenic epitopes region in protein.
Keywords
Ascariasis; Ascaris lumbricoides; Epitopes; Antigenic peptides; MHC-Binders; TapPred; PSSM; SVM; Nonamers; cytochrome c oxidase subunit 2 (mitochondrion)
Abbreviations
STH: soil-transmitted helminth; SAC: School-Age Children; MDA: Mass Drug Administration; MHC I: Major Histocompatibility Complex-Class I; MHC I: Major Histocompatibility Complex-Class II; PSSM: Position Specific Scoring Matrices; SVM: Support Vector Machine; GWD: Guinea Worm Disease; UniProt: The Universal Protein Resource; NCBI: National Center for Biotechnology Information; TAP: Transporter Associated with Antigen Processing; HPLC: High Performance Liquid Chromatography.
Introduction
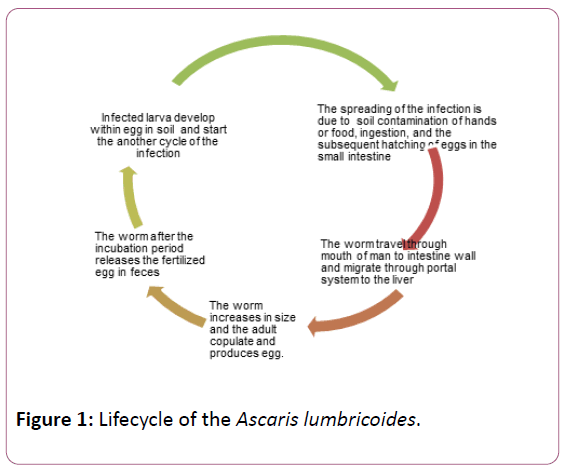
Ascaris lumbricoides is the causative agent of “Ascariasis” in human and other mammals by intestinal roundworm of the genus ‘Ascaris’. This parasite is the one of the largest nematode and, the measures of the adult worm measures upto 15-35 cm long and are white or yellow in color. the incubation period of the parasite is 10-24 months in the jejunum and middle ileum of the intestine( of host). Study suggest that, te infected individual sheds the different percentages of the eggs (fertilized egg: 45%, fertilized and unfertilized eggs shedding: 40% and unfertilized eggs: 20%. The fertilized eggs becomes infectious with period of 5-10 days and remain vible for upto 17 months after gets released in the favorable soil [1]. The spreading of the infection is due to soil contamination of hands or food, ingestion, and the subsequent hatching of eggs in the small intestine. The occurrence of this parasite was observed as a zoonotic infection which usually associated with pigs and use of hog manure [2] but, in most endemic areas, it is most likely transmitted from person to person [3].
The infected individuals, experience the symptoms of pneumonitis including wheezing, dyspnea, nonproductive cough, hemoptysis, and fever, and eosinophilia. The larvae incubation timing in the jejunum expected as 65 days and feeds on digestion of the host. The infected children with a marginal diet found to be susceptible to protein, caloric, or suffer from Vitamin A deficiency, which stood up in growth retardation and increases the susceptibility to malaria infection [4] and sometime in overexposed situation, it causes relative immunodeficiency [5]. In the higher infection load of this worm causes severe complication whereas, at minimal level it remains asymptomatic. The occurrence of this disease is worldwide but predominantly found in that geographical area with warm and moist climates especially common in tropical and subtropical areas where there is poor practice of the sanitation and hygiene. The estimated worldwide preponderance of this disease is 25% (0.8-1.22 billion people) [6]. Children’s of tropical and arising countries are more prone to this infection, where they are carried on by contamination of soil by human feces or use of untreated feces as fertilizer [7]. Symptomatic ascariasis may manifest as growth retardation, pneumonitis, intestinal obstruction, or hepatobiliary and pancreatic injury but, in most endemic areas, it is most likely transmitted from person to person [3]. The differential diagnoses are acute pancreatitis , biliary colic and community-acquired pneumonia. The vulnerable group for soil-transmitted helminth (STH) infections are children and as well as the impact of this infections directly on their health, nutritious status and cognitive ability [8]. The current strategy to control the STH infection is preventive chemotherapy with albendazole or mebendazole [9]. In the other study conducted by researcher in Chencha district where mass drug administration (MDA) to the most risky population including school-age children (SAC) is used as the central strategy to control soil-transmitted helminth (STH) infection [10]. In this study cytochrome c oxidase subunit II (mitochondrion) protein has been used to investigate its role in antigenicity. Cytochrome c oxidase subunit 2, also known as cytochrome c oxidase polypeptide II which is an oligomeric enzyme, an important component of the respiratory chain which involves in the transfer of electrons from cytochrome c to oxygen. This enzyme complex is found located in the mitochondrial inner membrane in eukaryotes. Cytochrome c oxidase subunit 2 contains two adjacent transmembrane regions in its Nterminus. The considerable part of the protein is generally exposed to the periplasmic or to the mitochondrial intermembrane space. The N-terminal domain of cytochrome C oxidase contains two transmembrane alpha-helices. Cytochrome oxidase deficiency and abnormality has been seen in the Leigh’s disease. Investigation shows that any alterations in the catalytic genes of cytochrome c oxidase subunits I and II (COI and COII) have an adverse impact on prognosis in patients with acute myeloid leukaemia (AML) [11]. A "mitochondrial hypothesis" of late onset Alzheimer's disease (AD) has been proposed. The in depth biochemical studies propose that there is a significant decrease in cytochrome oxidase (CO) activity as well as perturbed CO I and CO III mRNA levels in platelets and brain tissue from Alzheimer's patients. The phenotypic expression study of the CO mutation is the major reason for reduced CO activity and compromised mitochondrial function [12]. Antigen protein prediction from Ascaris l umbricoides is necessary for few paradigms of synthetic vaccine development and target validation. Antigenicity prediction of the protein from Ascaris lumbricoides can play an important role in prototype synthetic vaccine development and as well as for target validation.
Methodology
Retrieval of protein data from database
The protein amino acid sequence of cytochrome c oxidase subunit 2 (mitochondrion)from Ascaris lumbricoides parasite was retrieved from www.ncbi.nlm.nih.gov, UniProt databases [13-15].
Protein antigenicity prediction
Prediction of antigenicity Cytochrome c oxidase subunit 2 (mitochondrion) proteins were performed and obtained the peptide segments of protein that are likely to be antigenic and capable of eliciting an antibody response. The methods were used to analyze the antigenic peptide region of Cytochrome c oxidase subunit 2 (mitochondrion) are Hopp and Woods, Welling, Parker, Bepipred ,Kolaskar and Tongaonkar antigenicity methods[16-28].
MHC binding peptide prediction
The major histocompatibility complex (MHC) peptide binding of Ascarislumbricoidespredicted using neural networks trained on C terminals of known epitopes. Rankpep predicting tool predicts peptide binders to MHC-I ligands using PSSMs, whose C-terminal end is likely to be the result of proteosomal cleavage. The sequence similarity is observed to the peptides that bind to a given MHC molecule. Traditionally, the sequence patterns used for the prediction of peptides binding to MHC molecules. Such sequence patterns, however, have proven to be too simple, as the complexity of the binding motif cannot be precisely represented by the few residues present in the pattern [29]. RANKPEP uses “Position Specific Scoring Matrices (PSSMs) or profiles” from set of aligned peptides known to bind to a given MHC molecule as the predictor of MHC-peptide binding and overcome the complexity of the binding motif limitation. RANKPEP web server is a variability masking feature to focus on the prediction of conserved epitopes, which could thus help to avoid immune evasion resulting from mutation. Support Vector Machine (SVM) based method for prediction of promiscuous MHC class II binding peptides from protein sequence; SVM has been trained on the binary input of single amino acid sequence [30-33].
Prediction of antigenic peptides by cascade SVM based TAPPred method
In the present study, we predicted the cascade SVM based several TAP binders which was based on the sequence and the features of amino acids [34]. We found the MHCI binding regions (Table 3), the binding affinity of Ascaris lumbricoides.
Solvent accessible regions
We also analyzed the solvent accessible regions of proteins having highest probability that a given protein region lies on the surface of a protein Surface Accessibility, backbone or chain flexibility by Emini et al. [35] and Karplus and Schulz [36]. The different scaleswere used to predict the hydrophobic and hydrophilic characteristics of amino acids which is rich in charged and polar residues. The methods used are Sweet et al., Kyte and Doolittle, Abraham and Leo, Bull and Breese, Guy, Miyazawa, et al, Roseman, Wolfenden et al., Wilson et al., Cowan, Chothia [37-46].
Results
Retrieved protein information
The Ascaris lumbricoides antigenCytochrome c oxidase subunit 2 (mitochondrion)[gi|319656158|gb|ADV58572.1|], contain a long residue of 232 amino acids with 224 nonamers.
>gi|319656161|gb|ADV58575.1| cytochrome c oxidase subunit 2 (mitochondrion) [Ascarislumbricoides]
MNNFFQDFSLLFSSSLFSSYMDWFYNFNCSLLFGVLSFVSTMFVYL LLSSFYFKSKKIEYQFGELLCSVFPTLILVMQMVPSLSLLYYYGLMNLDSS LTVKVTGHQWYWSYEFSDIPGLEFDSYMKSLDQLELGEPRLLEVDNR CVVPCDVNIRFCITSGDVIHSWALPSMSIKLDAMSGILSTLSYSFPVVG VFYGQCSEICGANHSFMPVALEVTLLDNFKSWCMGLLND
Antigenic peptides result data through antigenic prediction method
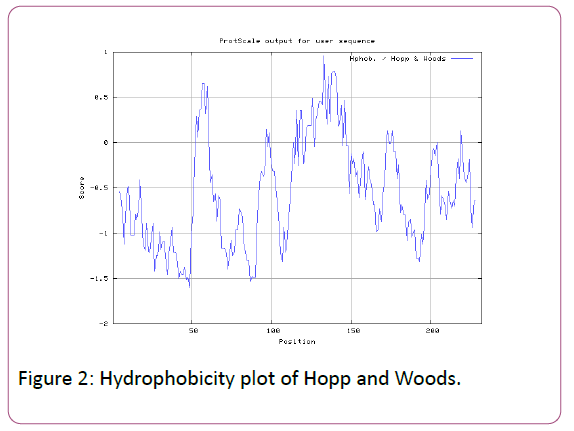
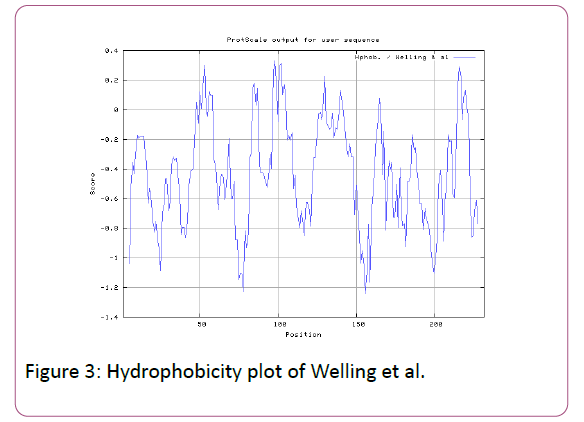
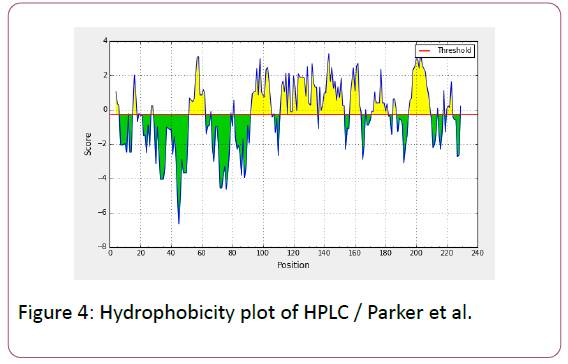
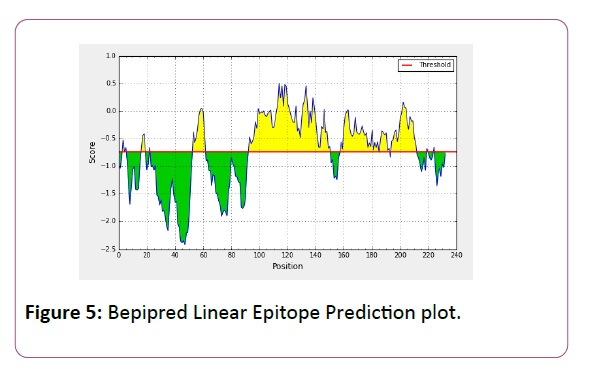
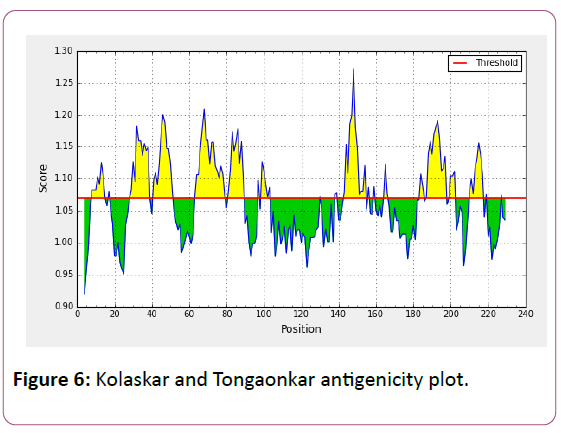
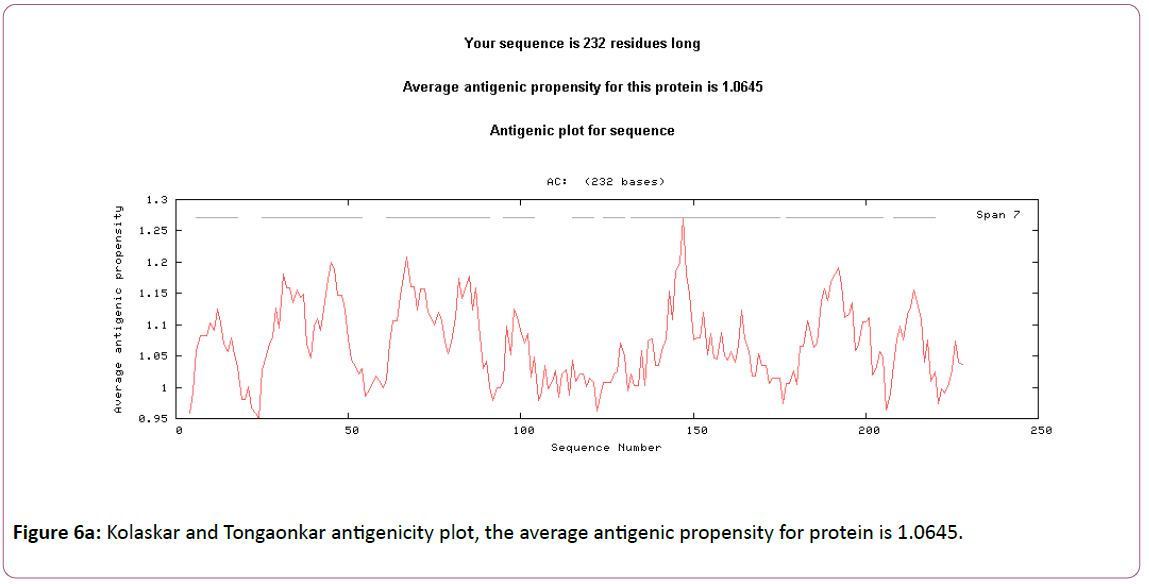
In this prediction, we investigated the area of greatest local Hydrophilicity through antigenic determinants . In the Hopp- Woods scale Hydrophilicity Prediction Result Data found high in Position: 133 Score: 0.956 (max) (130-QLELGEP -136) in a protein, assuming that the antigenic determinants would be exposed on the surface of the protein and thus would be located in hydrophilic regions (Figure 2). Welling et al. antigenicity plot provides value as the log of the quotient between percentage in average proteins and percentage in a sample of known antigenic regions. The prediction result found highest in Position: 98, Score: 0.331 (max) 95- DSSLTVK-101 (Figure 3). We also study Hydrophobicity plot of HPLC / Parker Hydrophilicity Prediction Result Data found in Position:143 (Residue:N) i.e. 140- EVDNRCV-146 with highest score:3.286 (Figure 4), BepiPred predicts the location of linear B-cell epitopes Result found in Position:114(Residue:S) with highest score :0.510 (111-YEFSDIP-117) (Figure 5), Kolaskar and Tongaonkar antigenicity methods (Figures 6a and 6b) Predicted peptides result found
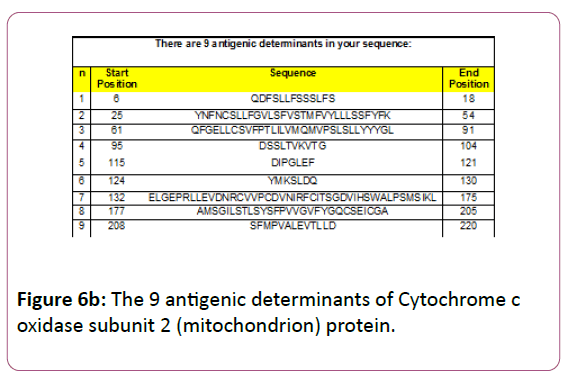
6-QDFSLLFSSSLFS-18,25- YNFNCSLLFGVLSFVSTMFVYLLLSSFYFK-54,61- QFGELLCSVFPTLILVMQMVPSLSLLYYYGL-91,95- DSSLTVKVTG-104, 115-DIPGLEF-121,124-YMKSLDQ-130,132 ELGEPRLLEVDNRCVVPCDVNIRFCITSGDVIHSWALPSMSIKL-175,1 77 AMSGILSTLSYSFPVVGVFYGQCSEICGA-205, 208- SFMPVALEVTLLD-220 and the predicted antigenic fragments can bind to MHC molecule is the first bottlenecks in vaccine design.
The predicted peptide of MHC class I & II
We found the binding of peptides to a number of different alleles using Position Specific Scoring Matrix. cytochrome c oxidase subunit 2 (mitochondrion) of Ascaris lumbricoides antigen, with sequence 232 amino acid residues long, having 224nonamers. MHC molecules are cell surface proteins, which actively participate in host immune reactions and involvement of MHC-I and MHC-II in response to almost all antigens. We have predicted MHC-I peptide binders of cytochrome c oxidase subunit 2 (mitochondrion) from Ascaris lumbricoides. We found predicted MHC-I peptide binders of protein for Matrix: 8mer_H2_Db.p.mtx, Consensus: QNWNCCTI, Optimal Score: 52.494, Binding Threshold: 33.04; Matrix: 9mer_H2_Db.p.mtx, Consensus: FCIHNCDYM, Optimal Score: 50.232, Binding Threshold: 17.96; Matrix: 10mer_H2_Db.p.mtx, Consensus: SGYYNFFWCL, Optimal Score: 58.858, Binding Threshold: 41.32;Matrix: 11mer_H2_Db.p.mtx,Consensus: CGVYNFYYCCY, Optimal Score: 79.495, Binding Threshold: 56.96 (Table 1) and MHC-II peptide binders for Matrix: I_Ab.p.mtx, Consensus: YYAPWCNNA, Optimal Score: 35.632, Binding Threshold: 9.52; Matrix: I_Ad.p.mtx, Consensus: QMVHAAHAE, Optimal Score: 53.145, Binding Threshold: 7.10; Matrix: I_Ag7.p.mtx, Consensus: WYAHAFKYV, Optimal Score: 40.873,Binding Threshold:7.54 for MHC II allele (Table 2) was tested. In this test, we found the MHCI and MHCII binding regions. T cell immune responses are derived by antigenic epitopes hence their identification is important for design synthetic peptide vaccine. T cell epitopes are recognized by MHCI molecules producing a strong defensive immune response against Ascaris lumbricoides antigen cytochrome c oxidase subunit 2. Therefore, the prediction of peptide binding to MHCI molecules by appropriate processing of antigen peptides occurs by their binding to the relevant MHC molecules. Because, the C-terminus of MHCI-restricted epitopes results from cleavage by the proteasome and thus, proteasome specificity is important for determining T-cell epitopes. Consequently, RANKPEP also focus on the prediction of conserved epitopes. C-terminus of MHCI-restricted peptides is generated by the proteasome, and thus RANKPEP also determines whether the C-terminus of the predicted MHCIpeptide binders is the result of proteasomal cleavage. Moreover, these sequences are highlighted in purple in the output results. Proteasomal cleavage predictions are carried out using three optional models obtained applying statistical language models to a set of known epitopes restricted by human MHCI molecules as indicated as I_Ab.p, I_Ad.p,I_Ag7.p,I_Ak.p alleles, which is highlighted in red represent predicted binders. Peptides whose score is above the binding threshold will appear highlighted in red and peptides produced by the cleavage prediction model are highlighted in violet. We also use a cascade SVM based TAPPred method which found 72 High affinity TAP Transporter peptide regions (Table 3) which represents predicted TAP binders residues which occur at N and C termini from Ascaris lumbricoides cytochrome c oxidase subunit 2.
Table 1: Promiscuous MHC ligands, having C-terminal ends are proteosomal cleavage sites of Ascaris lumbricoides. The antigenic peptide to the MHC-1 Allele i.e. 8mer_H2_Db(The binding thresholds: 33.04,optimal score: 52.494), 9mer_H2_Db(Optimal Score: 50.232, Binding Threshold: 17.96), 10mer_H2_Db(The Optimal Score: 58.858,BindingThreshold: 41.32), 11mer_H2_Db (Optimal Score: 79.495, Binding Threshold: 56.96). (All rows highlighted in red represent predicted binders & A peptide highlighted in violet has a C-teminus predicted by the cleavage model used).
| MHC-I Allele | RANK | POS. | N | SEQUENCE | C | MW (Da) | SCORE | % OPT. |
|---|---|---|---|---|---|---|---|---|
| 8mer_H2_Db | 1 | 151 | PCD | VNIRFCIT | SGD | 947.16 | 18.097 | 34.47% |
| 8mer_H2_Db | 2 | 114 | YEF | SDIPGLEF | DSY | 858.96 | 11.881 | 22.63% |
| 8mer_H2_Db | 3 | 81 | QMV | PSLSLLYY | YGL | 937.12 | 9.399 | 17.90% |
| 8mer_H2_Db | 4 | 140 | RLL | EVDNRCVV | PCD | 915.03 | 8.385 | 15.97% |
| 8mer_H2_Db | 5 | 39 | LSF | VSTMFVYL | LLS | 941.15 | 8.221 | 15.66% |
| 9mer_H2_Db | 1 | 24 | MDW | FYNFNCSLL | FGV | 1102.28 | 23.546 | 46.75% |
| 9mer_H2_Db | 2 | 202 | CSE | ICGANHSFM | PVA | 961.12 | 21.224 | 42.40% |
| 9mer_H2_Db | 3 | 155 | NIR | FCITSGDVI | HSW | 936.09 | 20.262 | 40.23% |
| 9mer_H2_Db | 4 | 145 | DNR | CVVPCDVNI | RFC | 943.14 | 18.155 | 36.05% |
| 9mer_H2_Db | 5 | 222 | LDN | FKSWCMGLL | ND | 1043.34 | 17.041 | 33.84% |
| 10mer_H2_Db | 1 | 217 | LEV | TLLDNFKSWC | MGL | 1185.39 | 20.603 | 35.00% |
| 10mer_H2_Db | 2 | 37 | GVL | SFVSTMFVYL | LLS | 1175.41 | 15.765 | 26.78% |
| 10mer_H2_Db | 3 | 89 | LYY | YGLMNLDSSL | TVK | 1094.25 | 12.727 | 21.62% |
| 10mer_H2_Db | 4 | 179 | DAM | SGILSTLSYS | FPV | 1009.13 | 11.842 | 20.12% |
| 10mer_H2_Db | 5 | 22 | SYM | DWFYNFNCSL | LFG | 1267.42 | 9.333 | 15.86% |
| 11mer_H2_Db | 1 | 217 | LEV | TLLDNFKSWCM | GLL | 1316.58 | 15.277 | 19.22% |
| 11mer_H2_Db | 2 | 33 | SLL | FGVLSFVSTMF | VYL | 1216.46 | 11.657 | 14.66% |
| 11mer_H2_Db | 3 | 190 | YSF | PVVGVFYGQCS | EIC | 1137.32 | 11.369 | 14.30% |
| 11mer_H2_Db | 4 | 22 | SYM | DWFYNFNCSLL | FGV | 1380.58 | 10.926 | 13.74% |
| 11mer_H2_Db | 5 | 81 | QMV | PSLSLLYYYGL | MNL | 1270.51 | 9.556 | 12.02% |
Table 2: Prediction of MHCII ligands all rows highlighted in red represent predicted binders to the MHC-II Allele i.e. MHC-II I_Ab, MHC-II I_Ad,MHC-II I_Ag7. (All rows highlighted in red represent predicted binders).
| MHC-II Allele | RANK | POS. | N | SEQUENCE | C | MW (Da) | SCORE | % OPT. |
|---|---|---|---|---|---|---|---|---|
| MHC-II I_Ab | 1 | 24 | MDW | FYNFNCSLL | FGV | 1102.28 | 15.46 | 43.39% |
| MHC-II I_Ab | 2 | 146 | NRC | VVPCDVNIR | FCI | 996.19 | 11.457 | 32.15% |
| MHC-II I_Ab | 3 | 166 | IHS | WALPSMSIK | LDA | 991.25 | 9.113 | 25.58% |
| MHC-II I_Ab | 4 | 86 | LSL | LYYYGLMNL | DSS | 1131.36 | 8.789 | 24.67% |
| MHC-II I_Ab | 5 | 66 | GEL | LCSVFPTLI | LVM | 974.23 | 8.484 | 23.81% |
| MHC-II I_Ad | 1 | 179 | DAM | SGILSTLSY | SFP | 922.05 | 11.052 | 20.80% |
| MHC-II I_Ad | 2 | 6 | NFF | QDFSLLFSS | SLF | 1025.14 | 10.975 | 20.65% |
| MHC-II I_Ad | 3 | 56 | FKS | KKIEYQFGE | LLC | 1123.28 | 10.568 | 19.89% |
| MHC-II I_Ad | 4 | 198 | FYG | QCSEICGAN | HSF | 906 | 9.117 | 17.15% |
| MHC-II I_Ad | 5 | 33 | SLL | FGVLSFVST | MFV | 938.09 | 7.251 | 13.64% |
| MHC-II Ag7 | 1 | 114 | YEF | SDIPGLEFD | SYM | 974.05 | 7.637 | 18.68% |
| MHC-II Ag7 | 2 | 214 | PVA | LEVTLLDNF | KSW | 1045.2 | 6.26 | 15.32% |
| MHC-II Ag7 | 3 | 190 | YSF | PVVGVFYGQ | CSE | 947.1 | 5.899 | 14.43% |
| MHC-II Ag7 | 4 | 78 | LVM | QMVPSLSLL | YYY | 969.21 | 4.86 | 11.89% |
| MHC-II Ag7 | 5 | 81 | QMV | PSLSLLYYY | GLM | 1100.3 | 4.423 | 10.82% |
Table 3: Cascade SVM based High affinity TAP Binders of Ascaris lumbricoides.
| Peptide Rank | Start Position | Sequence | Score | Predicted Affinity |
|---|---|---|---|---|
| 1 | 94 | LDSSLTVKV | 8.645 | High |
| 2 | 34 | GVLSFVSTM | 8.641 | High |
| 3 | 6 | QDFSLLFSS | 8.64 | High |
| 4 | 163 | IHSWALPSM | 8.639 | High |
| 5 | 55 | SKKIEYQFG | 8.637 | High |
| 6 | 13 | SSSLFSSYM | 8.635 | High |
| 7 | 147 | VPCDVNIRF | 8.63 | High |
| 8 | 82 | SLSLLYYYG | 8.627 | High |
| 9 | 215 | EVTLLDNFK | 8.627 | High |
| 10 | 185 | LSYSFPVVG | 8.619 | High |
Prediction of solvent accessible regions of protein
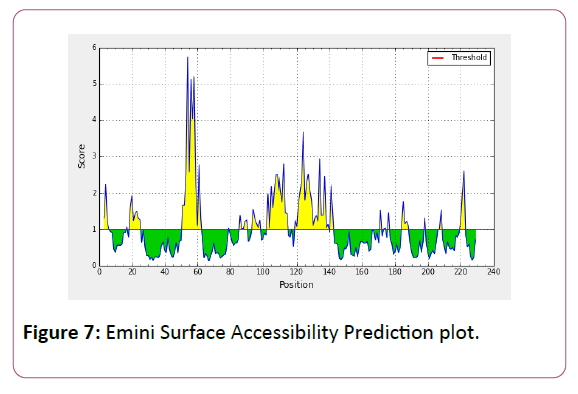
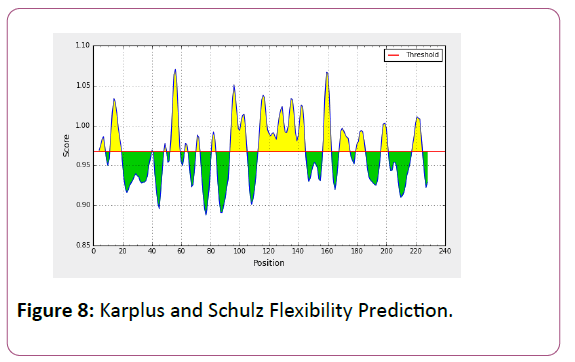
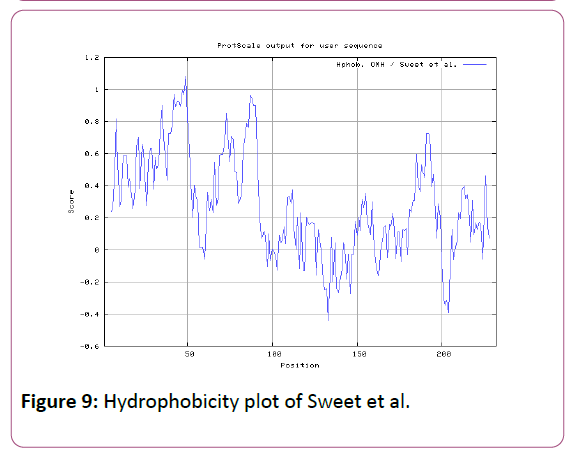
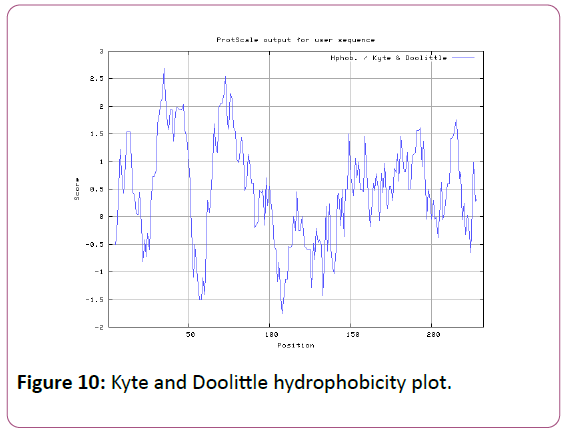
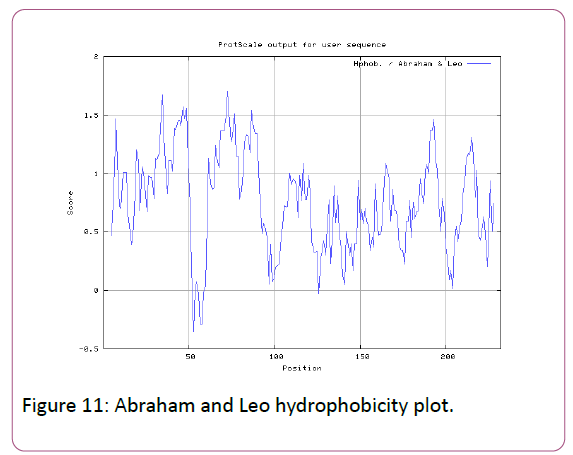
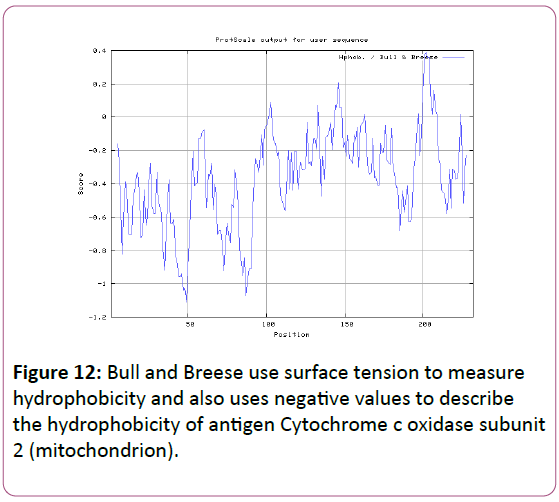
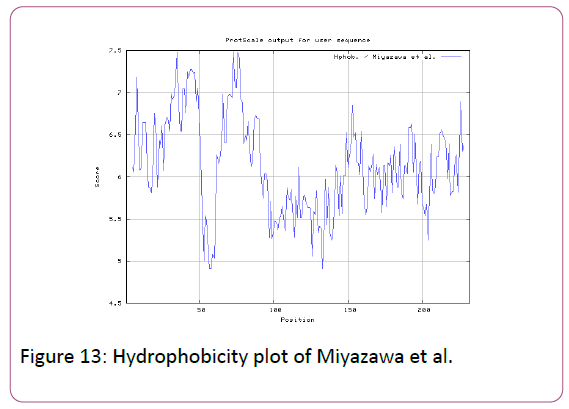
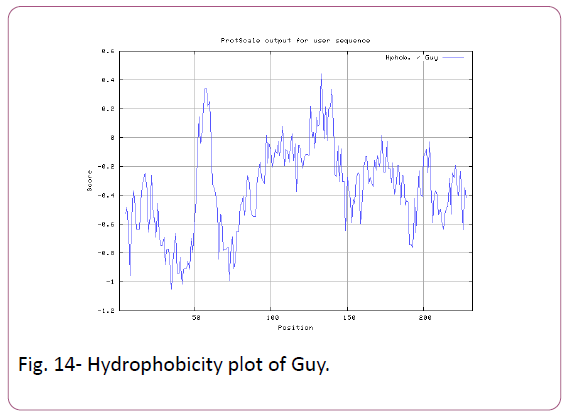
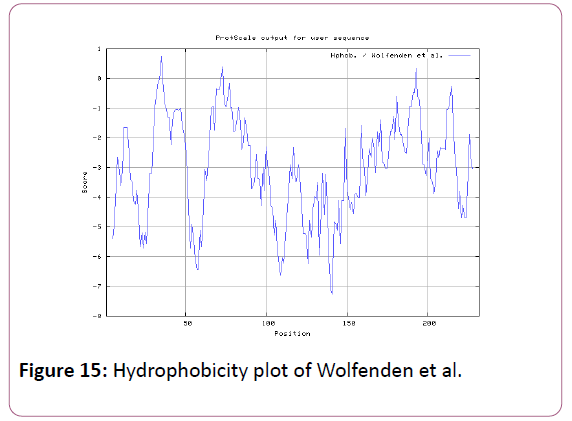
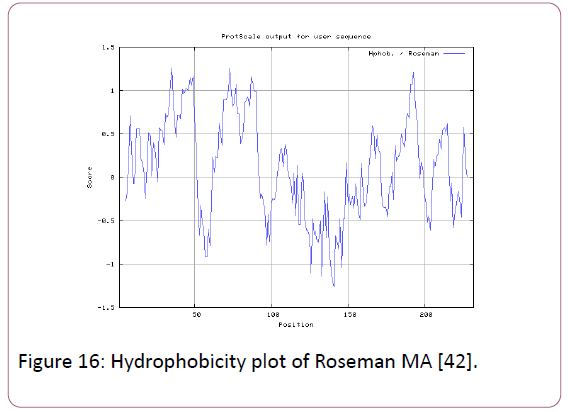
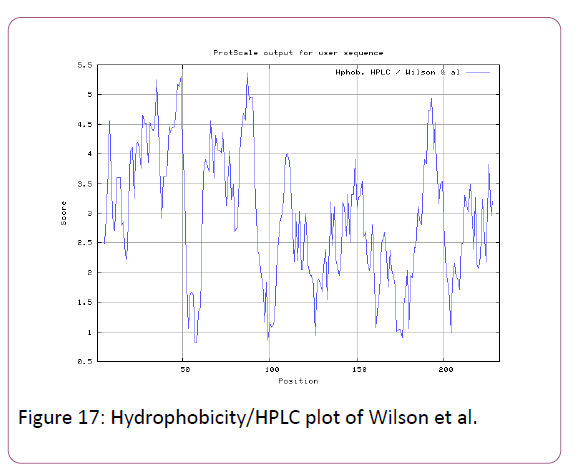
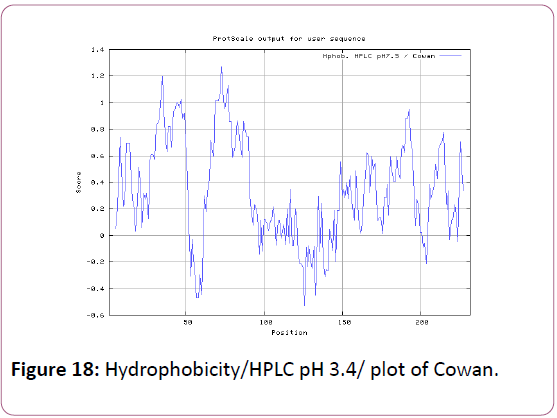
We also predict solvent accessible regions in proteins; different measurement was performed for the prediction of antigenic activity, surface region of peptides. Emini et al., (Figure 7) predicts the highest probability in position:54 (Residue: K) i.e., 52-YFKSKK-57 with maximum score:5.745, that a given protein region lies on the surface of a protein and are used to identify antigenic determinants on the surface of proteins. Karplus and Schulz (Figure 8) High score is found i.e. found in position:56 (Residue:K) 53-FKSKKIE-59 with maximum score: 1.071. The hydrophobicity and hydrophilic characteristics of amino acids is determined by several other scales i.e. Sweet et al. hydrophobicity prediction result data found high in position in Position:49 with maximum Score: 1.082 (max)(46-LLLSSFY-52) (Figure 9), Kyte & Doolittle result high in position:35, Score:2.689 (max)(32-LFGVLSF-38) (Figure 10), Abraham and Leo result high Position:73, Score:1.707 (max)(70-FPTLILV-76) (Figure 11), Bull and Breese use surface tension to measure in Position: 202, Score:0.387 (max)(199- CSEICGA-205) (Figure 12), Miyazawa result high in Position:73 Score:7.486 (max)(70-FPTLILV-76 (Figure 13), Guy result high in Position:133 Score:0.444 (max)(130-QLELGEP-136) (Figure 14), Wolfenden result high in Position:35, Score:0.737 (max)(32- LFGVLSF-38) (Figure 15), Roseman result high in Position: 35,Score:1.262 (max)(32-LFGVLSF-38) (Figure 16), Wilson et al. Position:87, Score:5.367 (max)(84-SLLYYYG-90) (Figure 17), Cowan Position: 73, Score: 1.270 (max)(70-FPTLILV-76) (Figure 18), ChothiaPosition: 35, Score: 0.446 (max)(32- LFGVLSF-38) (Figure 19).
Conclusion
MHC molecules are the cell surface proteins, which actively take part in the host immune responses against infection (pathogens) and reason of its involvement in the response to almost all antigens and it gives effects on specific sites. This knowledge of the immune responses to an antigen protein (cytochrome c oxidase subunit 2 from Ascaris lumbricoides) clear that the whole protein is not necessary for raising the immune response, but a small fragment of antigen can induce immune response against whole antigen. This means the increase in affinity of MHC binding peptides may result in enhancement of immunogenicity of cytochrome c oxidase subunit 2 from Ascaris lumbricoides, hence are helpful in silico to design and develop highly predictive computational tools for the identification of T-cell epitopes. Finally, accurate prediction remains vital for the future to design synthetic peptide vaccine. The Overall conducted study and opted results are encouraging. Both the ‘sites of action’ and ‘physiological functions’ can be predicted with very high accuracies which is helping to minimize the number of validation experiments. The future perspectives of this method will be useful in cellular immunology, vaccine design, immunodiagnostics, immunotherapeutic and molecular understanding of autoimmune susceptibility.
References
- Peng W, Zhou X, Gasser RB (2003) Ascaris egg profiles in human faeces: biological and epidemiological implications. Parasitology 127: 283-290.
- Hoenigl M, Seeber K, Valentin T, Zollner-Schwetz I, Krause R (2010-2013) Pulmonary ascariasis in patients from wealthy countries: shift in epidemiology?. Int J Infect Dis. 2012 Dec. 16(12):e888;Notes from the field: ascariasis associated with pig farming - Maine, MMWR Morb Mortal Wkly Rep. 2013 62:413.
- Peng W, Yuan K, Hu M, Gasser RB (2007) Recent insights into the epidemiology and genetics of Ascaris in China using molecular tools. Parasitology 134: 325-330.
- Le Hesran JY, Akiana J, Ndiaye el HM, Dia M, Senghor P, et al. (2004) Severe malaria attack is associated with high prevalence of Ascarislumbricoides infection among children in rural Senegal. Trans R Soc Trop Med Hyg 98: 397-399.
- Dold C, Holland CV (2011) Ascarisand ascariasis. Microbes Infect 13:632-637.
- Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, et al. (2006) Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet 367: 1521-1532.
- Walker M, Hall A, Basáñez MG (2011) Individual predisposition, household clustering and risk factors for human infection with Ascarislumbricoides: new epidemiological insights. PLoSNegl Trop Dis 5:e1047.
- Zhou Q, Liu CF, Zhang LX, Zhou H, Chen YD (2015) [Research progress in soil-transmitted helminth infection control among children at home and abroad]. ZhongguoXue Xi Chong Bing Fang ZhiZaZhi 27: 431-435.
- Speich B, Moser W, Ali SM, Ame SM, Albonico M, et al. (2016) Efficacy and reinfection with soil-transmitted helminths 18-weeks post-treatment with albendazole-ivermectin, albendazole-mebendazole, albendazole-oxantelpamoate and mebendazole. Parasit Vectors 9: 123.
- Zerdo Z, Yohanes T, Tariku B (2016) Soil-Transmitted Helminth Reinfection and Associated Risk Factors among School-Age Children in Chencha District, Southern Ethiopia: A Cross-Sectional Study. J Parasitol Res 4737891.
- Silkjaer T, Nyvold CG, Juhl-Christensen C, Hokland P, Nørgaard JM (2013) Mitochondrial cytochrome c oxidase subunit II variations predict adverse prognosis in cytogenetically normal acute myeloid leukaemia. Eur J Haematol 91:295-303.
- Hamblet NS, Ragland B, Ali M, Conyers B, Castora FJ (2006) Mutations in mitochondrial-encoded cytochrome c oxidase subunits I, II, and III genes detected in Alzheimer's disease using single-strand conformation polymorphism. Electrophoresi 27:398-408
- https://www.ncbi.nlm.nih.gov
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, et al. (2012). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 40(Database issue): D13-D25.
- Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, et al. (2005) The Universal Protein Resource (UniProt). Nucleic Acids Res 33: D154-159.
- Hopp TP, Woods KR (1981) Prediction of protein antigenic determinants from amino acid sequences. ProcNatlAcadSci U S A 78: 3824-3828.
- Welling GW, Weijer WJ, van der Zee R, Welling-Wester S (1985) Prediction of sequential antigenic regions in proteins. FEBS Lett 188: 215-218.
- Parker KC, Bednarek MA, Coligan JE (1994) Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol 152: 163-175.
- Larsen JE, Lund O, Nielsen M (2006) Improved method for predicting linear B-cell epitopes. Immunome Res 2: 2.
- Kolaskar AS, Tongaonkar PC (1990) A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 276: 172-174.
- Gomase VS, Kale KV, Chikhale NJ, Changbhale SS (2007) Prediction of MHC binding peptides and epitopes from alfalfa mosaic virus. Curr Drug DiscovTechnol 4: 117-215.
- Gomase VS,KaleKV (2008) In silico prediction of epitopes: a new approach for fragment based viral peptide vaccines. Int J of Applied Computing 1: 39-46.
- Gomase VS, Kale KV (2008) Approach of proteomics system architecture in plant virus’s database". Int. J. of Applied Computing 1: 33-38.
- Gomase VS, Chitlange NR (2012) Sensitive Quantitative Predictions of MHC Binding Peptides and Fragment Based Peptide Vaccines from Taeniacrassiceps. J Vaccines Vaccin3:131.
- Gomase VS, Chitlange NR (2012) Microbial Proteomics Approach for Sensitive Quantitative Predictions of MHC Binding Peptide from Taeniaovis. J Data Mining Genomics Proteomics 3:121.
- Mishra Sonu, Virendra SGomase (2015) Prediction of antigenic epitope from D. medinensis: new paradigm of synthetic vaccine development. International Conference on “Recent Research Development in Environment, Social Sciences and Humanities” (ICRRDESH-15) :103-107.
- Mishra Sonu, Virendra S. Gomase (2015) Analysis of hydrophobicity and antigenic epitope prediction from D. Medinensis. International Conference on “Technologies for Sustainability-Engineering, information Technology,Management and the Environment”(SUSTECH-15) : 44-51.
- Mishra SonuandVirendra S. Gomase (2015). Analysis Of Hydrophobicity And Antigenicity Of Heat Shock Protein 70 From GWD. 2ndInternational Conference on “Recent Innovations in science, Engineering and Management”(ICRISEM-15), pp-25-33. ISBN: 978-81-931039-9-9.
- Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, et al. (1993) Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell 74: 929-937.
- Reche PA, Glutting JP, Reinherz EL (2002) Prediction of MHC class I binding peptides using profile motifs. Hum Immunol 63: 701-709.
- Reche PA, Reinherz EL (2003) Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J MolBiol 331: 623-641.
- Craiu A, Akopian T, Goldberg A, Rock KL (1997) Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. ProcNatlAcadSci U S A 94: 10850-10855.
- Pieters J (2000) MHC class II-restricted antigen processing and presentation. AdvImmunol 75: 159-208.
- Bhasin M, Raghava GP (2004) Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci 13: 596-607.
- Emini, E.A., Hughes, J.V., Perlow, D.S., Boger, J (1985). Induction of hepatitis a virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 55: 836-839.
- Karplus PA, Schulz GE. (1985). Prediction of chain flexibility in proteins: a tool for the selection of peptide antigen. Naturwissenschaften. 72: 212-213.
- Sweet RM, Eisenberg D (1983) Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J MolBiol 171: 479-488.
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J MolBiol 157: 105-132.
- Abraham DJ, Leo AJ (1987) Extension of the fragment method to calculate amino acid zwitterion and side chain partition coefficients. Proteins 2: 130-152.
- Bull HB, Breese K (1974) Surface tension of amino acid solutions: a hydrophobicity scale of the amino acid residues. Arch BiochemBiophys 161: 665-670.
- Miyazawa S, Jernigen RL (1985) Estimation of Effective Interresidue Contact Energies from Protein Crystal Structures: Quasi-Chemical Approximation. Macromolecules 18: 534-552.
- Roseman MA (1988). Hydrophilicity of polar amino acid side-chains is markedly reduced by flanking peptide bonds. J MolBiol 200: 513-522.
- Wolfenden R, Andersson L, Cullis PM, Southgate CC (1981) Affinities of amino acid side chains for solvent water. Biochemistry 20: 849-855.
- Wilson KJ, Honegger A, Stötzel RP, Hughes GJ (1981) The behaviour of peptides on reverse-phase supports during high-pressure liquid chromatography. Biochem J 199: 31-41.
- Cowan R, Whittaker RG (1990) Hydrophobicity indices for amino acid residues as determined by high-performance liquid chromatography. Pept Res 3: 75-80.
- Chothia C (1976) The nature of the accessible and buried surfaces in proteins. J MolBiol 105: 1-12.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences