Gender Differences in the Expression of Proteins in Autoreactive T-cells Specific to Central Nervous System Myelin Proteolipid Protein 139-151
Chandirasegaran Massilamany, Renu Nandakumar, Nandakumar Madayiputhiya, Sanjit Pandey, Chittibabu Guda and Jay Reddy
Chandirasegarsan Massilamany1, Renu Nandakumar1, Nandakumar Madayiputhiya1, Sanjit Pandey2, Chittibabu Guda2, and Jay Reddy3*
1Nebraska Redox Biology Centre, University of Nebraska-Lincoln, Lincoln, NE, 68588, USA
2University of Nebraska Medical Centre, Omaha, NE 68198, USA
3School of Veterinary Medicine and Biomedical Sciences, University of Nebraska-Lincoln, Lincoln, NE 68583, USA
- *Corresponding Author:
- Jay Reddy
School of Veterinary Medicine and Biomedical Sciences
University of Nebraska-Lincoln, Lincoln
NE 68583, USA
Tel: +14024728541
E-mail: nreddy2@unl.edu
Received date: November 24, 2016; Accepted date: January 06, 2017; Published date: January 13, 2017
Citation: Massilamany C, Nandakumar R, Madayiputhiya N, Pandey S, Guda C, et al. (2016) Gender Differences in the Expression of Proteins in Autoreactive T-cells Specific to Central Nervous System Myelin Proteolipid Protein 139-151. J Autoimmune Disord 3:1. doi: 10.21767/2471-8513.100030
Copyright: © 2017 Massilamany C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Autoimmune diseases are more prevalent in women than in men, and such a disparity also exists in animal models. In our studies with the mouse model of experimental autoimmune encephalomyelitis (EAE) induced with myelin proteolipid protein 139-151 (PLP) for multiple sclerosis, gender differences are well documented, in that only female, but not male, SJL mice show chronic relapsingremitting paralysis. Since EAE is typically mediated by CD4 T cells, we tested a hypothesis that the generation and expansion of antigen-specific CD4 T cells might be different between genders.
Methods and findings: First, by creating a newer and more sensitive version of major histocompatibility complex class II tetramers called dextramers, we noted no differences in the generation of PLP-specific T cells in the peripheral repertoires of male and female SJL mice. Second, comparable numbers of PLP-specific T cells were found to be infiltrated into the brains of both genders as verified by flow cytometric analysis. Third, evaluation of molecules that positively or negatively regulate the expansion of effector T cells revealed that expression of cytotoxic T-lymphocyte associated protein 4 was tended to be more in the T cells that infiltrate into the brains of male than female mice. Finally, we analyzed the proteomic profiles in PLP 139-151- specific T cells to identify additional novel molecules that may potentially contribute to the gender-disparity in the development of central nervous system autoimmunity. In conjunction with tandem mass spectrometric analysis and ingenuity pathway analysis, we have identified a panel of proteins found to be expressed gender dependently, suggesting that they can be regulated by sex steroids.
Conclusions: Our data revealed expression of proteins that can potentially play a role in the development of diseases of multiple organs such as the central nervous system and cardiovascular system including cancers.
Keywords
Gender Differences; Central Nervous System; Multiple Sclerosis; Systemic Lupus Erythematosus
Abbreviations
EAE: Experimental Autoimmune Encephalomyelitis; PLP: Myelin Proteolipid Protein; MS: Multiple Sclerosis; SLE; Systemic Lupus Erythematosus; DCM: Dilated Cardiomyopathy; DHT: Dihydrotestosterone; IL: Interleukin; IFN: Interferon; ESR: Estrogen Receptor; MHC: Major Histocompatibility Complex; CTLA-4: Cytotoxic T-lymphocyte Associated Protein 4; CNS: Central Nervous System; CFA: Complete Freund’s Adjuvant; NASE: Neuraminidase; 3[H]: tritiated; LNC: Lymph Node Cells; MNC: Mononuclear Cells; 7-AAD: 7-amino-actinomycin-D; ICOS: Inducible T-cell Costimulator; PD-1: Programmed Cell Death Protein-1; AR: Androgen Receptor; MS/MS: Tandem Mass Spectrometric Analysis; ERBB2: Receptor Tyrosine-protein Kinase erbB2-precursor; NRIF-2: Neurotrophin Receptor-Interacting Factor 2; SPEN: MSX-2-interacting Protein; RTN2: Isoform 1 of Reticulon-2.
Introduction
Most autoimmune diseases like rheumatoid arthritis, systemic lupus erythematosus (SLE), and multiple sclerosis (MS) are more frequently seen in females than in males [1-3]. Gender differences also have been noted in cancer susceptibility. For example, the incidence of lung cancer in people who have never smoked is higher in women than men; liver cancer is more prevalent in men than women [4,5]. However, dilated cardiomyopathy (DCM) and myocarditis, including amyotrophic lateral sclerosis (>1.6:1), occur more frequently in men than women [6,7]. Likewise, hypertension, atherosclerosis, and stroke are lower in premenopausal women than in both men of similar age and postmenopausal women [6,8]. Specifically, the finding that women are more likely than men to develop MS suggests that the disease might be influenced by sex hormones [9]. While the disease severity in female MS patients is reduced during pregnancy, MS symptoms return to their pre-pregnant state within six months post-delivery [3,10,11]. Reduced severity of MS during pregnancy is correlated with elevated estriol levels, and oral estriol treatment of relapsing-remitting MS patients reduces enhancing lesions on magnetic resonance imaging [12]. All of these observations provide a compelling reason to study the role of sex hormones in modulating the pathogeneses and outcomes of inflammatory, immune-mediated, organ-specific autoimmune and metabolic diseases and cancers.
Our studies involve experimental autoimmune encephalomyelitis (EAE), the disease model being widely used to study MS pathogenesis and also in drug discovery research [13-15]. Among various EAE models, gender differences have been well documented with myelin proteolipid protein (PLP) 139-151-induced EAE in SJL mice [16,17]. While females show chronic relapsing-remitting paralysis, male mice show only the monophasic form [17]. However, clinical relapses could be induced in orchidectomized males, suggesting that androgens regulate antigen-specific T-cell expansion [16]. Additionally, estrogens and androgens have been shown to influence the severity of EAE [18-23]. For example, testosterone treatment of female mice was previously shown to augment the production of an anti-inflammatory cytokine, interleukin (IL)-10 [24]. However, our recent data with dihydrotestosterone (DHT) indicated no such deviation; rather DHT treatment of antigenprimed T cells in vitro led to cell death occurring in association with autophagy [25]. Conversely, ovariectomized mice treated with estradiol showed increased production of interferon (IFN)-γ [26]. Thus, the notion has evolved that sex steroids can act on T cells and modulate cytokine production [18]. In support of this hypothesis, splenocytes and thymocytes of mice and humans have been shown to express receptors for estrogens and androgens [27-29], and estrogen receptor (ESR)-responsive elements have been identified in IFN-γ and ESR genes [30,31]. Other potential mechanisms that may contribute to genderdisparity in the susceptibility to EAE include under production of IL-12 in male antigen presenting cells [32], variations in regulatory T cell functions [33] and sex chromosomes [34,35].
Since PLP 139-151-induced EAE is typically mediated by CD4 T cells, we hypothesized that gender differences, are expected to be contributed by this subset of T cells. In this report, by using major histocompatibility complex (MHC) class II/PLP 139-151 dextramers, we show that differences in T cell expansion are unlikely contributing factors for gender-biased EAE phenotypes, pointing to the role of additional unknown factors in the causation of disease. Our data however revealed that expression of cytotoxic T-lymphocyte associated protein 4 (CTLA-4) in the T cells infiltrating into the brains of male mice tended to be more than in female mice. Furthermore, by performing total cell proteomic analysis in T cells specific to PLP 139-151, we identified a panel of proteins expressed gender dependently, suggesting that they can be potentially regulated by sex steroids. Some of these proteins may have a role in the development of diseases of multiple organs such as the central nervous system (CNS) and cardiovascular system including cancers.
Materials and Methods
Mice and ethics statement
Male and female SJL/J (H-2s) mice (five-to-six-week-old) were obtained from the Jackson Laboratory (Bar Harbor, ME). The mice were maintained in accordance with the animal protocol guidelines of the University of Nebraska-Lincoln, Lincoln, NE. The study was conducted in accordance with the National Institutes of Health guidelines for the use of experimental animals, and the protocols were specifically approved by the University of Nebraska-Lincoln Institutional Animal Care and Use Committee (permit number: A3459-01; protocol #999).
Peptide synthesis and immunization procedures
The peptides PLP 139-151 (HSLGKWLGHPDKF) and neuraminidase (NASE) 101-120 (EALVRQGLAKVAYVYKPNNT) were synthesized based on 9-fluorenylmethyloxycarbonyl chemistry (Neopeptide, Cambridge, MA). All peptides were HPLC-purified (>90%), identity confirmed by mass spectroscopy, and dissolved in 1x PBS prior to use. To measure recall responses, peptides were emulsified in complete Freund’s adjuvant (CFA) and administered subcutaneously in multiple sites in the flank and sternum (100 μg/mouse). Additionally, for EAE-induction, pertussis toxin (100 mg/mouse) was administered on day 0 and day 2 [13,14].
T cell proliferation assay
Mice were killed ten days post immunization, and the draining lymph nodes were harvested to obtain single cell suspensions. Lymph node cells (LNC) were stimulated with PLP 139-151 and NASE 101-120 (control) at a cell density of 5 × 106 cells/ml in RPMI medium supplemented with 10% fetal calf serum, 1 mM sodium pyruvate, 4 mM L-glutamine, 1x each of non-essential amino acids and vitamin mixture, and 100 U/ml penicillinstreptomycin (Lonza). After two days, the cultures were then pulsed with 1 μCi of tritiated 3[H] thymidine per well; 16 h later, proliferative responses were measured as counts per minute (cpm) using the Wallac liquid scintillation counter (Perkin Elmer, Waltham, MA).
IAs dextramer staining
LNC cultures: To determine the frequency of antigen-specific T cells, IAs/PLP 139-151, and Theiler’s murine encephalomyelitis virus (TMEV), 70-86 dextramers were created as described [36]. LNC obtained from the PLP 139-151-immunized mice were stimulated with the peptide (20 μg/ml) for six days. Viable lymphoblasts were incubated with the previously standardized concentration of dextramers at room temperature for two hours [36]. Cells were washed, and stained with anti-CD4 and 7-aminoactinomycin- D (7-AAD; Invitrogen, Eugene, OR). After acquiring the cells by flow cytometry (FACS Calibur, BD Bioscience, San Diego, CA), frequency of dextramer+cells were determined using Flow Jo software (Tree Star, Ashland, OR) in the live (7-AAD-) CD4+ population.
Mononuclear cells (MNCs) from brains and spinal cords: EAE mice showing paralytic signs were perfused into the hearts as above. Brains were harvested by blunt dissection, and the spinal cords were flushed out using 1x cold HBSS. After homogenization and digestion with type IV collagenase (400 U/ml; Worthington, Lakewood, NJ) at 37°C for 1 h, MNCs were harvested by percoll density gradient centrifugation (70%/30%) as described previously [37]. Cells were then stained with IAs dextramers (PLP 139-151 or TMEV 70-86) followed by anti-CD4 and 7-AAD, and after cells were acquired by flow cytometry, dextramer+cells were determined as described above.
Surface staining for T-cell markers
MNCs harvested from EAE mice were stained with CD40L, inducible T-cell costimulator (ICOS), CTLA-4 and programmed cell death protein (PD)-1 antibodies or isotype controls (Armenian hamster IgG; all from eBioscience, San Diego, CA), and anti-CD4 and 7-AAD. After washing, cells were acquired by flow cytometry and the percentages of cells positive for each marker were analyzed in the live (7-AAD-) CD4+ population.
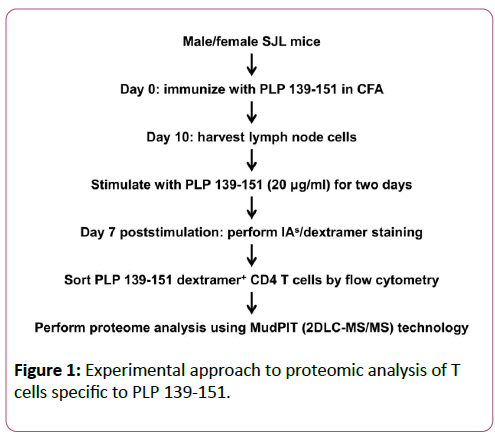
Protein sample preparation
The steps involved in the preparation of protein samples are schematically illustrated in Figure 1. LNC obtained from PLP 139-151-immunized male or female SJL mice were re-stimulated with the immunizing peptide (20 μg/ml) for two days. IL-2 medium was then supplemented, and on day 7, cells were stained with IAs/PLP 139-151 dextramers or IAs/TMEV 70-86 (control) dextramers, anti-CD4, and 7-AAD. PLP dextramer +CD4+live (7-AAD-) cells (3 × 105) were then sorted by flow cytometry (FACS Aria III, BD Biosciences) in two individual experiments. After pelleting, cells were lysed using radio immunoprecipitation assay buffer supplemented with a cocktail of protease inhibitors (Thermoscientific, Rockford, IL). Lysis was performed on ice over an orbital shaker for 30 min and then centrifuged at 14,000 rpm. After supernatants were harvested, proteins were precipitated by adding nine volumes of chilled acetone at -20°C overnight. The protein pellets were then obtained by centrifugation and subjected to in-solution trypsin digestion. Proteins were reduced using 10 mM dithiothreitol, alkylated with 40 mM iodoacetamide, and digested with trypsin (1:50 trypsin: protein ratio; Roche) overnight at 37°C. The tryptic peptides were then desalted and concentrated using PepClean C-18 spin columns according to the manufacturer’s instructions (Thermo Scientific).
Tandem mass spectrometric analysis (MS/MS) analysis
Fully automated two-dimensional chromatographic experiments were performed on protein samples using an ultimate 3000 Dionex multidimensional liquid chromatography (MDLC) system (Dionex Corporation) integrated with a nanospray source and LCQ Fleet Ion Trap mass spectrometer (Thermo Scientific). First-dimension liquid chromatography (LC) separation [Strong Cation-exchange (SCX) chromatography] with fraction collection was performed, followed by seconddimension LC separation (reverse phase chromatography) and peptide detection by tandem mass spectrometry [38,39]. The first-dimensional separation was performed on a SCX column (Polysulfoethyl, 1 mm I.D × 15 cm, 5 μm, 300A, Dionex). Samples (20 μl) were loaded onto the first-dimension SCX column and eluted using a salt gradient (0-600 mM) for 45 min. Based on the UV absorbance of the eluted peptides, selected fractions were subjected to second-dimension analysis. The second-dimension separation included on-line sample pre-concentration and desalting using a monolithic C 18 trap column (Pep Map, 300 μm I.D × 5 mm, 5 μm, 100A, Dionex). The loading of the sample on the monolithic trap column was done at a flow rate of 40 ml/ min. The desalted peptides were then eluted and separated on a C 18 Pep Map, 75 μm I.D × 15 cm, 3 μm, 100A column by applying an acetonitrile gradient (ACN plus 0.1% formic acid, 90- minute gradient at a flow rate of 200 nl/min) and introduced into the mass spectrometer using the nanospray source. The LCQ Fleet mass spectrometer was operated with the following parameters: nanospray voltage, 2.0 kV; heated capillary temperature, 200°C; and full scan m/z range, 400-2000. The LCQ was operated in data-dependent mode with 4 MS/MS spectra for every full scan, 5 microscan averaged for full scans and MS/MS scans, 3 m/z isolation width for MS/MS isolations, and 35% collision energy for collision-induced dissociation.
The MS/MS spectra were searched against the IPI mouse (version 3.26) database using MASCOT (Version 2.2 Matrix Science). Database search criteria were as follows: enzyme, Trypsin; missed cleavages, 2; mass, monoisotropic; fixed modification, carbamidomethyl (C); peptide tolerance, 1.5Da; MS/MS fragment ion tolerance, 1Da. Probability assessment of peptide assignments and protein identifications were accomplished using Scaffold software (version Scaffold 3.4, Proteome Software Inc.). Criteria for protein identification included detection of at least 2 uniquely identified peptides and a protein probability score of ≥ 90%. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Relative quantitation of the proteins was done based on the label-free method of spectral counting using the normalized spectral counts for each protein. A ≥ 2-fold change (p ≤ 0.05) in abundance was used to select for proteins that were differentially expressed between the male and female samples. A heat map of protein abundance was generated using log2 transformed normalized spectral counts of the proteins present in both male and female samples. Hierarchical cluster analysis was performed using cluster 3.0 by complete linkage clustering and the Pearson correlation (uncentered) similarity metric method; the heat map was visualized using Java Treeview.
Ingenuity pathway analysis (IPA)
The proteins that were expressed uniquely in males or females through MS/MS analysis were subjected to the IPA knowledge base to generate the networks for males (androgen receptor (AR)/testosterone) and females (ESR1/estrogen or ESR2/estrogen). In addition, a crisscross analysis was performed by subjecting the proteins that were expressed uniquely in males or females with ESR1/estrogen and ESR2/estrogen or AR/ testosterone, respectively.
Statistical analysis
Differences in T cell proliferation and marker+ cells between groups were analyzed by students.
t. P ≤ 0.05 were considered significant.
Results and Discussion
Gender disparity with females developing autoimmune diseases more frequently than males have been well documented, but the underlying molecular mechanisms continue to be enigmatic. In our studies with PLP 139-151- induced EAE, only females, but not males, show chronic relapsing-remitting disease [13,14]. Furthermore, PLP-induced EAE is typically mediated by CD4 T cells, and we have previously shown that EAE severity in male recipients of female PLP 139-151-specific T cells is comparable to severity in female recipients of female cells [14,16]. Conversely, female recipients of male PLP-specific T cells show less severe disease [14]. These findings suggest that the encephalitogenic potential of auto reactive T cells might be influenced by gender. Since CD4 T cells are indispensable mediators of EAE in SJL mice, we recently created MHC class II dextramers, more sensitive reagents than tetramers for PLP 139-151 that permitted us to determine gender-differences in the generation and expansion of T cell repertoires specific to PLP 139-151 [36].
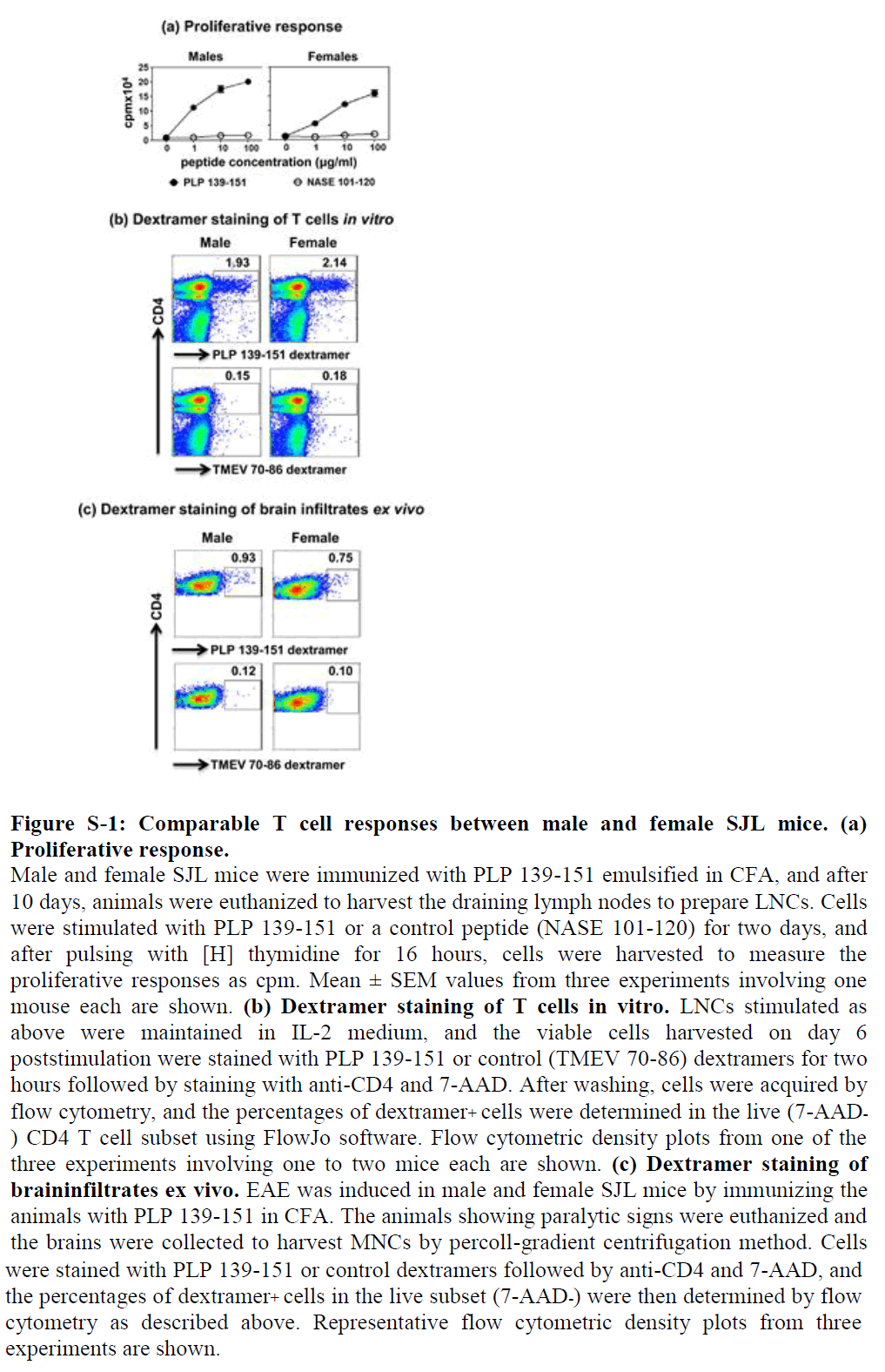
We evaluated PLP-specific T cell repertoires in three levels. We first immunized groups of male and female SJL mice, and after 10 days, prepared LNCs from the draining lymph nodes to ascertain recall responses to PLP 139-151. In a T cell proliferation assay, it was clear that cells from both genders responded equally to PLP 139-151, and the responses were antigen specific, since the cells did not respond to a control antigen (NASE 101-120) (Figure S1a). To further verify antigen specificity, we used IAs dextramers for PLP 139-151 (specific) and TMEV 70-86 (control), expecting that the PLP, but not TMEV, dextramers would bind PLP-responsive T cells. Of note, both PLP 139-151 and TMEV 70-86 peptides bind the same MHC class II allele, IAs; thus, TMEV 70-86 serves as an appropriate control antigen for PLP 139-151 [36]. Briefly, LNC obtained from immunized animals were stimulated with PLP 139-151 as described above, and the cells were stained with dextramers on day 6 post stimulation. The dextramer analysis revealed that about 2% of total CD4 T cells were found to be positive for PLP 139-151, whereas staining for control dextramers was negligible, and the proportions of antigen-specific T cells were similar for both genders (Figure S1b).
These data complemented the results obtained from the T cell proliferation assay as described above, leading us to conclude that antigen-specific T cells expand similarly in both genders. However, a possibility existed that dissimilar proportions of antigen-specific T cells may infiltrate into the brains. To address this question, we immunized groups of male and female SJL mice, and, as the animals developed paralysis, we harvested brain tissue to isolate MNCs as previously described [36].
By using flow cytometry, we estimated the frequencies of PLPspecific CD4 T cells using PLP 139-151 dextramers but found no significant differences between genders (males, 0.93% vs. females, 0.75%; Figure S1c), suggesting that antigen-specific T cells have easy access to the brain and are expected to mediate CNS damage similarly in both genders. This is the case, as both male and female SJL mice develop acute EAE with equal severity as we have previously described in an adoptive transfer-EAE protocol [14], but they differ in the chronic phase in that only females but not males show a chronic relapsing-remitting paralysis [16].
These findings may imply that the T cell responses might be regulated differentially between genders leading us to reason that the molecules implicated in the regulation of T cell responses might vary between genders. To address this question, we evaluated the expression of positive (CD40L and ICOS) and negative (CTLA-4 and PD-1) regulators using the LNCs and MNCs derived from PLP 139-151-immunized mice. We noted that the expression of these molecules were found not to vary in the PLP 139-151-sensitized CD4 T cells generated in the peripheral repertoires of male and female mice (data not shown). However, evaluation of these markers in the MNCs derived from brains revealed that only CTLA-4 was tended to be upregulated in males as compared to females (11.20 ± 1.22 % vs. 5.17 ± 1.92%, p=0.057; Table 1) Since, CTLA-4 has been identified as an important negative regulator of autoreactive T cells [40], its increased expression in males may mean that the expansion of antigen-specific T cells may be inhibited leading to their quicker recovery than females.
| Markers | Males | Females |
|---|---|---|
| CD40L | 3.70 ± 2.11 | 1.05 ± 0.45 |
| ICOS | 81.19 ± 5.84 | 67.7 ± 10.08 |
| CTLA-4* | 11.20 ± 1.22 | 5.17 ± 1.92 |
| PD-1 | 37.7 ± 3.49 | 30.59± 5.02 |
| Ham IgG | 0.57± 0.17 | 0.91 ± 0.20 |
| Numbers represent mean ± SEM values (n=3); Hamster IgG, control for CD40L, CTLA-4, PD-1, ICOS; *p=0.057 | ||
Table 1: Percentages of CD4 T cells expressing various cell surface markers in the brain MNCs purified from EAE mice.
Next, we sought to identify novel molecules in the PLPspecific T cells that may contribute to gender variations in their EAE phenotypes. To this end, we used PLP 139-151 dextramers to sort PLP-specific T cells by flow cytometry, and performed proteomic analysis on dextramer+ cells purified from male and female mice. We sorted 3 × 105 CD4+PLP 139-151 dextramer+ cells with a purity of 100% in two individual experiments.
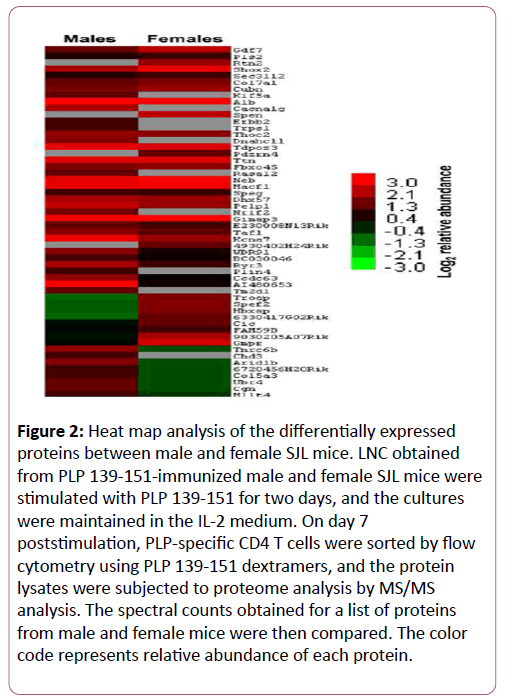
The protein lysates were subjected to proteomic analysis to identify differentially expressed proteins using MASCOT and Scaffold bioinformatics software. The analyses revealed differential expression of 54 proteins (Figure 2 and Table 2). Criteria for protein identification included detection of at least two uniquely identified peptides for the protein in question with a probability score of ≥ 90%. A ≥ 2-fold change (p ≤ 0.05) in abundance was used to select differentially expressed proteins (Figure 2 and Table 2), yielding identities of 40 proteins. Among these, a panel of proteins was found to be uniquely expressed either in males (10) or females (4) or upregulated in males (11) or females (9). Most of these proteins have been linked to various disease conditions including cancer and autoimmunity.
| Name of the protein | Accession # | Gene | Fold change |
|---|---|---|---|
| Uniquely expressed in males | |||
| Kinesin heavy chain isoform 5A | IPI00109420 | Kif5a | 2.2 |
| Neurotrophin receptor-interacting factor 2 | IPI00121598 | Nrif2/ZNF274 | 2.7 |
| Calcium channel, voltage-dependent, T type, alpha 1G subunit | IPI00123380 | Cacna1g | 4 |
| RAS protein activator like 2 | IPI00405756 | Rasal2 | 2.2 |
| Left-right dynein | IPI00622122 | Dnahc11 | 3.1 |
| Similar to beta-amyloid binding protein precursor | IPI00753190 | Tm2d1 | 2.2 |
| Similar to zinc finger transcription factor TRPS1 | IPI00749846 | Trps1 | 1.8 |
| Chromodomain helicase DNA binding protein 3 | IPI00551435 | Chd3 | 1.8 |
| Receptor tyrosine-protein kinase erbB-2 precursor | IPI00626433 | Erbb2 | 1.8 |
| S3-12 | IPI00131113 | Plin4 | 1.8 |
| Uniquely expressed in females | |||
| Isoform 1 of Reticulon-2 | IPI00120427 | Rtn2 | 3.4 |
| Isoform 1 of Msx2-interacting protein | IPI00378158 | Spen | 4.5 |
| Similar to PDZ domain-containing RING finger protein 4 | IPI00752560 | Pdzrn4 | 2.3 |
| Novel protein | IPI00757104 | 4930402H24Rik | 2.3 |
| Upregulated in males | |||
| Trinucleotide repeat containing 6b isoform 1 | IPI00226311 | Tnrc6b | 5.9 |
| WD repeat-containing protein 91 | IPI00339693 | WDR91 | 2.6 |
| Zinc finger, UBR1 type 1 | IPI00378681 | Ubr4 | 4.2 |
| Isoform 1 of uncharacterized protein C14orf101 homolog | IPI00409254 | 6720456H20Rik | 3.4 |
| Ryanodine receptor 3 | IPI00606638 | Ryr3 | 2.4 |
| Collagen type V alpha 3 chain | IPI00626350 | Col5a3 | 3.4 |
| Similar to Afadin | IPI00753111 | Mllt4 | 3 |
| Similar to AT rich interactive domain 1B (SWI1-like) isoform 3 | IPI00756022 | Arid1b | 5.8 |
| Similar to Cingulin | IPI00756207 | Cgn | 4.2 |
| Hypothetical protein LOC330188 | IPI00808379 | Ccdc63 | 3 |
| Unregulated in females | |||
| Isoform 2 of growth/differentiation factor 7 precursor | IPI00118190 | Gdf7 | 2 |
| Isoform 1 of short stature homeobox protein 2 | IPI00230342 | Shox2 | 2 |
| Trophinin associated protein | IPI00406255 | TROAP | 6.7 |
| KPL2 protein | IPI00458397 | Spef2 | 6 |
| Isoform 1 of protein capicua homolog | IPI00653910 | Cic | 2.5 |
| Similar to remodeling and spacing factor 1 isoform 12 | IPI00676351 | Hbxap | 6 |
| Similar to guanosine monophosphate reductase isoform 2 | IPI00753090 | Gmpr | 5.8 |
| Similar to gamma-aminobutyric acid (GABA-B) receptor binding protein | IPI00755658 | 6330417G02Rik | 5.3 |
| Similar to CG5237-PA | IPI00757967 | BC030046 | 4.4 |
Table 2: Differentially expressed proteins in PLP 139-151-specific CD4 T cells derived from male and female SJL mice.
Figure 2: Heat map analysis of the differentially expressed proteins between male and female SJL mice. LNC obtained from PLP 139-151-immunized male and female SJL mice were stimulated with PLP 139-151 for two days, and the cultures were maintained in the IL-2 medium. On day 7 poststimulation, PLP-specific CD4 T cells were sorted by flow cytometry using PLP 139-151 dextramers, and the protein lysates were subjected to proteome analysis by MS/MS analysis. The spectral counts obtained for a list of proteins from male and female mice were then compared. The color code represents relative abundance of each protein.
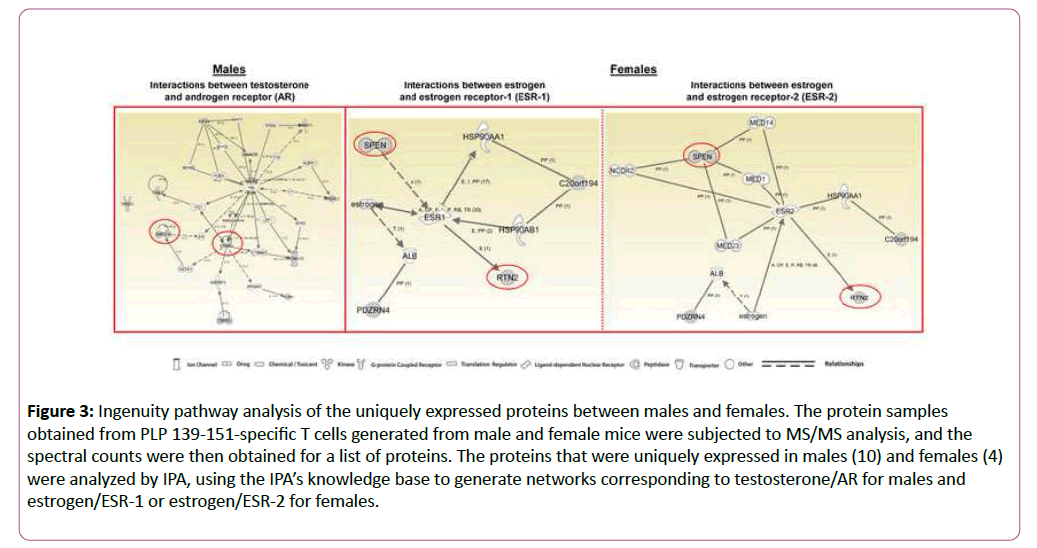
To determine whether the proteins expressed uniquely in males and females can be regulated by their corresponding hormones and receptors, androgens/AR and estrogens/ESR-1 and ESR-2, we used IPA. The network generated using ten proteins expressed uniquely in males (Table 2) led us to identify several intermediary proteins that directly and/or indirectly interact with AR and testosterone (Figure 3, left panel). Important among these are receptor tyrosine-protein kinase erbB2-precursor (ERBB2) and neurotrophin receptor-interacting factor 2 (Nrif2 designated, ZNF274); ERBB2 is expected to interact with four proteins uniquely identified in males including ZNF274 (Table 2 and Figure 3, left panel). While ERBB2, a wellknown breast cancer gene, also is required for development of oligodendrocytes [41], Nrif2/ZNF274 plays a role in cell cycle progression, programmed cell death, neuronal cholesterol synthesis, and a number of neurodegenerative diseases [42,43].
Figure 3: Ingenuity pathway analysis of the uniquely expressed proteins between males and females. The protein samples obtained from PLP 139-151-specific T cells generated from male and female mice were subjected to MS/MS analysis, and the spectral counts were then obtained for a list of proteins. The proteins that were uniquely expressed in males (10) and females (4) were analyzed by IPA, using the IPA’s knowledge base to generate networks corresponding to testosterone/AR for males and estrogen/ESR-1 or estrogen/ESR-2 for females.
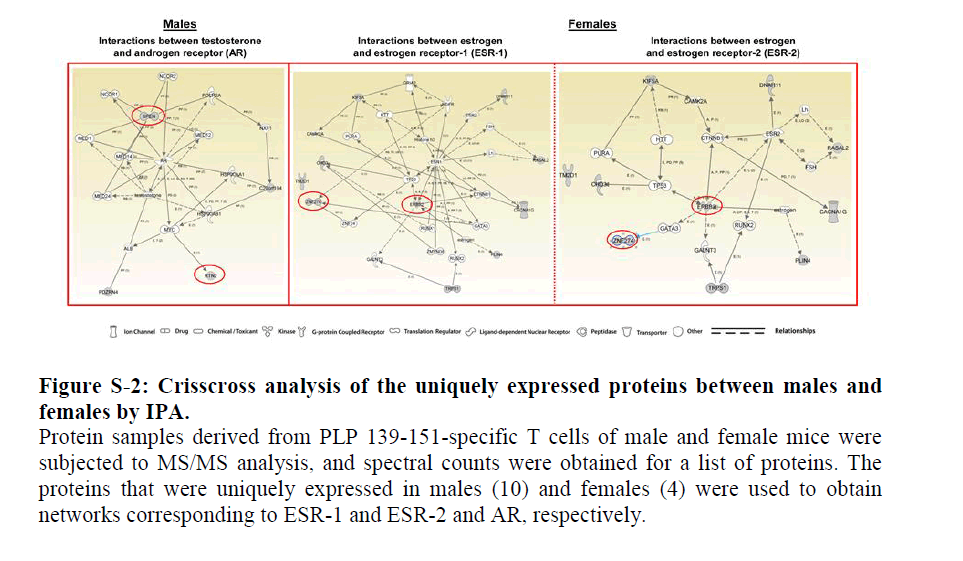
In a crisscross analysis, we then asked whether the proteins uniquely expressed in males can also interact with ESRs. The patterns of interactions for the above two proteins were essentially similar with both ESR-1 and ESR-2 (Figure S2, middle and right panels). In a similar analysis, we generated a network using the four uniquely expressed proteins in females (Table 2) corresponding to ESR-1 and ESR-2, leading us to identify two proteins, isoform 1 of MSX-2-interacting protein (SPEN) and isoform 1 of reticulon-2 (RTN2), that interacted with both receptors (Figure 3, middle and right panels). While RTN-2 showed a direct interaction with both ESR-1 and ESR-2, interaction of SPEN was indirect requiring mediator complex subunits as intermediaries. Involvement of such intermediaries also was noted with AR in a crisscross analysis (Figure S2, left panel).
Biologically, RTN-2 is implicated in neurodegeneration, and SPEN can promote transcriptional activation in osteoblasts and act as a negative regulator of the Notch pathway in addition to blocking the differentiation of B cells in the marginal zone and thymocytes [44-48].
Overall, the data suggest that the proteins expressed uniquely in males and females may carry out functions under the influence of their corresponding sex steroids, testosterone in males and estrogens in females. However, their functionalities also can be potentially influenced by the hormones of the opposite sex.
Gender differences have been noted in a number of autoimmune diseases, cardiomyopathy, cancers, and stroke [1-3,6,49,50]. Furthermore, autoimmune diseases (e.g., SLE, Sjogren’s syndrome) and cancers (e.g., non-Hodgkin’s lymphoma, Hodgkin’s disease) can coexist [51-55], suggesting that common pathogenetic mechanisms support the development of both conditions. Likewise, while most autoimmune diseases commonly occur in women, men are more likely than women to develop cardiovascular diseases or strokes, suggesting a role for sex hormones to influence disease outcomes. However, a direct causal link between sex hormones and predisposition to cardiovascular diseases continues to be clinically tenuous. Furthermore, clinical outcomes of hormone replacement therapy have been variable [56-58], in part because steroid hormones can affect numerous cell types and modulate a multitude of gene expressions by acting as transcriptional factors [59,60]. Importantly, steroid receptors also can be activated by ligands and non-ligands [61,62], making it difficult to define and predict the benefits or side effects of steroid hormones. One way to alleviate or minimize side effects is through derivation of small chemical molecules as modulators of AR and ESR functions [61,63]. Alternatively, identification of molecules downstream of sex hormone signalling events provides avenues to use them as therapeutic targets. Pursuit of the latter option led us to identify proteins that may play a gender-dependent role. Important among these are two uniquely expressed proteins in males –ErbB2, which is a breast cancer gene that has been implicated in cardiomyopathies and myocardial ischemia and also in cardiomyocyte development [64-67]; and Nrif2/ZNF274, a transcriptional factor shown to be associated with cell death and neurodegenerative diseases [43,68] and one protein expressed uniquely in females –SPEN, a transcriptional factor that regulates the Notch pathway and differentiation of immune cells [69].
In summary, we have identified a panel of proteins that could be involved in the downstream events or signalling pathways of sex steroids, androgens in males and estrogen and its derivatives in females. Autoimmune diseases and cancers share a common feature to some degree with respect to immune recognition of antigenic determinants. While self-antigens become targets for immune attack in autoimmune diseases, immune cells recognize altered self-antigens in cancers [70,71]. In our studies, we used a unique system in that SJL mice are prone to both tumour development (sarcomas) and autoimmunity (PLP-induced EAE) [72,73]. Therefore, the proteins that we have identified may have a role in the development of diseases of multiple organs such as CNS and cardiovascular system including cancers. Alternatively, common pathways may exist for their occurrence and our data provide avenues to delineate such possibilities.
Conflict of interest
None declared.
References
- Lockshin MD (2001) Invited review: sex ratio and rheumatic disease. J ApplPhysiol91:2366-73.
- Ahmed SA, Hissong BD, Verthelyi D, Donner K, Becker K (1999) Gender and risk of autoimmune diseases: possible role of estrogenic compounds. Environ Health Perspect Suppl 5:681-686.
- Whitacre CC (2001) Sex differences in autoimmune disease. Nat Immunol 2:777-80.
- Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, et al. (2007) Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 317:121-124.
- Gazdar AF, Thun MJ (2007) Lung cancer, smoke exposure, and sex. J Clin Oncol 25:469-471.
- Rosamond W, Flegal K, Furie K, Go A, Greenlund K, et al. (2008) Heart disease and stroke statistics update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 117: 121-126.
- Czlonkowska A, Ciesielska A, Gromadzka G, Kurkowska-Jastrzebska I (2006) Gender differences in neurological disease: role of estrogens and cytokines. Endocrine 29:243-256.
- Turtzo LC, McCullough LD (2008) Sex differences in stroke. Cerebrovasc Dis 26:462-474.
- Harbo HF, Gold R, Tintore M (2013) Sex and gender issues in multiple sclerosis. Therapeutic advances in neurological disorders 6:237-248.
- Confavreux C, Hutchinson M, Hours MM, Cortinovis-Tourniaire P, Moreau T (1998) Rate of pregnancy-related relapse in multiple sclerosis Pregnancy in Multiple Sclerosis Group. N Engl J Med 339:285-291.
- Lockshin MD (2006) Sex differences in autoimmune disease. Orthop Clin North Am 37:629-633.
- Soldan SS, Alvarez Retuerto AI, Sicotte NL, Voskuhl RR (2003) Immune modulation in multiple sclerosis patients treated with the pregnancy hormone estriol. J Immunol 171:6267-674.
- Massilamany C, Steffen D, Reddy J (2010) An epitope from Acanthamoebacastellanii that cross-react with proteolipid protein 139-151-reactive T cells induces autoimmune encephalomyelitis in SJL mice. J Neuroimmunol 219:17-24.
- Massilamany C, Thulasingam S, Steffen D, Reddy J (2011) Gender differences in CNS autoimmunity induced by mimicry epitope for PLP 139-151 in SJL mice. J Neuroimmunol230:95-104.
- Miller SDK (2007) Experimental autoimmune encephalomyelitis in the mouse. Current Protocols in Immunol ogy Inc 15: 19-23.
- Bebo BF, Schuster JC, Vandenbark AA, Offner H (1998) Gender differences in experimental autoimmune encephalomyelitis develop during the induction of the immune response to encephalitogenic peptides. J Neurosci Res 52:420-426.
- Bebo BF, Vandenbark AA, Offner H (1996) Male SJL mice do not relapse after induction of EAE with PLP 139-151. J Neurosci Res45:680-689.
- Bebo BF, Fyfe-Johnson A, Adlard K, Beam AG, Vandenbark AA (2001) Low-dose estrogen therapy ameliorates experimental autoimmune encephalomyelitis in two different inbred mouse strains. J Immunol 166:2080-2089.
- Bebo BF, Schuster JC, Vandenbark AA, Offner H (1999) Androgens alter the cytokine profile and reduce encephalitogenicity of myelin-reactive T cells. J Immunol 162:35-40.
- Palaszynski KM, Liu H, Loo KK, Voskuhl RR (2004) Estriol treatment ameliorates disease in males with experimental autoimmune encephalomyelitis: implications for multiple sclerosis. J Neuroimmunol 149:84-89.
- Offner H (2004) Neuroimmunoprotective effects of estrogen and derivatives in experimental autoimmune encephalomyelitis: therapeutic implications for multiple sclerosis. J Neurosci Res 78:603-624.
- Garidou L, Laffont S, Douin-Echinard V, Coureau C, Krust A (2004) Estrogen receptor alpha signaling in inflammatory leukocytes is dispensable for 17beta-estradiol-mediated inhibition of experimental autoimmune encephalomyelitis. J Immunol 173:2435-2442.
- Lelu K, Laffont S, Delpy L, Paulet PE, Perinat T, et al. (2011)Estrogen receptor alpha signaling in T lymphocytes is required for estradiol-mediated inhibition of Th1 and Th17 cell differentiation and protection against experimental autoimmune encephalomyelitis. J Immunol 187:2386-2893.
- Dalal M, Kim S, Voskuhl RR (1997) Testosterone therapy ameliorates experimental autoimmune encephalomyelitis and induces a T helper 2 bias in the autoantigen-specific T lymphocyte response. J Immunol 159:3-6.
- Jia T, Anandhan A, Massilamany C, Rajasekaran RA, Franco R (2015) Association of Autophagy in the Cell Death Mediated by Dihydrotestosterone in Autoreactive T Cells Independent of Antigenic Stimulation. J Neuroimmune Pharmacol10:620-634.
- Maret A, Coudert JD, Garidou L, Foucras G, Gourdy P, et al. (2003) Estradiol enhances primary antigen-specific CD4 T cell responses and Th1 development in vivo. Essential role of estrogen receptor alpha expression in hematopoietic cells. Eur J Immunol 33:512-521.
- Benten WP, Becker A, Schmitt-Wrede HP, Wunderlich F (2002) Developmental regulation of intracellular and surface androgen receptors in T cells. Steroids67:925-31.
- Benten WP, Lieberherr M, Giese G, Wrehlke C, Stamm O, et al. (1999) Functional testosterone receptors in plasma membranes of T cells. Faseb J13:123-133.
- Kovacs WJ, Olsen NJ (1987) Androgen receptors in human thymocytes. J Immunol 139:490-493.
- Fox HS, Bond BL, Parslow TG (1991)Estrogen regulates the IFN-gamma promoter. J Immunol 146:4362-4367.
- Kube D, Platzer C, von Knethen A, Straub H, Bohlen H, et al. (1995) Isolation of the human interleukin 10 promoter. Characterization of the promoter activity in Burkitt's lymphoma cell lines. Cytokine7:1-7.
- Kim S, Voskuhl RR (1999) Decreased IL-12 production underlies the decreased ability of male lymph node cells to induce experimental autoimmune encephalomyelitis. J Immunol 162:5561-5568.
- Reddy J, Illes Z, Zhang X, Encinas J, Pyrdol J, et al. (2004) Myelin proteolipid protein-specific CD4+CD25+ regulatory cells mediate genetic resistance to experimental autoimmune encephalomyelitis. Proc Natl Acad Sci 101:15434-1549.
- Sasidhar MV, Itoh N, Gold SM, Lawson GW, Voskuhl RR (2012) The XX sex chromosome complement in mice is associated with increased spontaneous lupus compared with XY. Ann Rheum Dis 71:1418-1422.
- Smith-Bouvier DL, Divekar AA, Sasidhar M, Du S, Tiwari-Woodruff SK, et al. (2008) A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med 205:1099-1108.
- Massilamany C, Upadhyaya B, Gangaplara A, Kuszynski C, Reddy J (2011) Detection of autoreactive CD4 T cells using major histocompatibility complex class II dextramers. BMC Immunol 12:40.
- Korn T, Reddy J, Gao W, Bettelli E, Awasthi A, et al. (2007) Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat Med 13:423-431.
- Du Z, Nandakumar R, Nickerson KW, Li X (2015) Proteomic adaptations to starvation prepare Escherichia coli for disinfection tolerance. Water Res 69:110-119.
- Fernandez J, Marroquin-Guzman M, Nandakumar R, Shijo S, Cornwell KM, et al.(2014) Plant defence suppression is mediated by a fungal sirtuin during rice infection by Magnaportheoryzae. Mol Microbiol 94:70-88.
- McCoy KD, Le Gros G (1999) The role of CTLA-4 in the regulation of T cell immune responses. Immunol Cell Bio l77:1-10.
- Park SK, Miller R, Krane I, Vartanian T (2001) The erbB2 gene is required for the development of terminally differentiated spinal cord oligodendrocytes. J Cell Biol154:1245-1258.
- Korade Z, Kenchappa RS, Mirnics K, Carter BD (2009) NRIF is a regulator of neuronal cholesterol biosynthesis genes. J Mol Neurosci 38:152-158.
- Kruttgen A, Saxena S, Evangelopoulos ME, Weis J. Neurotrophins and neurodegenerative diseases: receptors stuck in traffic? J Neuropathol Exp Neurol 62:340-350.
- Kuroda K, Han H, Tani S, Tanigaki K, Tun T (2003) Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18:301-312.
- Montenegro G, Rebelo AP, Connell J, Allison R, Babalini C, et al.(2012) Mutations in the ER-shaping protein reticulon 2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J Clin Invest122:538-544.
- Newberry EP, Latifi T, Towler DA (1999) The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry 38:10678-10690.
- Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T (2007) Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc Natl Acad Sci 104:1610-1615.
- VanderWielen BD, Yuan Z, Friedmann DR, Kovall RA (2011) Transcriptional repression in the Notch pathway: thermodynamic characterization of CSL-MINT (Msx2-interacting nuclear target protein) complexes. J Biol Chem 286:14892-14902.
- Beeson PB (1994) Age and sex associations of 40 autoimmune diseases. Am J Med 96:457-462.
- Tang MX, Jacobs D, Stern Y, Marder K, Schofield P, et al. (1996) Effect of oestrogen during menopause on risk and age at onset of Alzheimer's disease. Lancet 348:429-432.
- Lechner K, Chen YA (2010)Paraneoplastic autoimmune cytopenias in Hodgkin lymphoma. Leuk Lymphoma 51:469-474.
- Martin DN, Mikhail IS, Landgren O (2009) Autoimmunity and hematologic malignancies: associations and mechanisms. Leuk Lymphoma 50:541-550.
- Javierre BM, Esteller M, Ballestar E (2008) Epigenetic connections between autoimmune disorders and haematological malignancies. Trends Immunol 29:616-623.
- Abu-Shakra M, Ehrenfeld M, Shoenfeld Y (2002) Systemic lupus erythematosus and cancer: associated or not? Lupus 11:137-144.
- Kiss E, Kovacs L, Szodoray P (2010) Malignancies in systemic lupus erythematosus. Autoimmun Rev 9:195-199.
- Anderson GL, Limacher M, Assaf AR, Bassford T, Beresford SA, et al. (2004) Effects of conjugated equine estrogen in postmenopausal women with hysterectomy: the Women's Health Initiative randomized controlled trial. JAMA 291:1701-1712.
- Siegel C, Turtzo C, McCullough LD (2010) Sex differences in cerebral ischemia: possible molecular mechanisms. J Neurosci Res88:2765-2774.
- Wassertheil-Smoller S, Hendrix SL, Limacher M, Heiss G, Kooperberg C, et al. (2003) Effect of estrogen plus progestin on stroke in postmenopausal women: the Women's Health Initiative: a randomized trial. JAMA 289:2673-2684.
- Klein-Hitpass L, Schwerk C, Kahmann S, Vassen L (1998) Targets of activated steroid hormone receptors: basal transcription factors and receptor interacting proteins. J Mol Med (Berl) 76:490-496.
- Falkenstein E, Tillmann HC, Christ M, Feuring M, Wehling M (2000) Multiple actions of steroid hormones--a focus on rapid, nongenomic effects. Pharmacol Rev 52:513-516.
- Clarke BL, Khosla S (2010) Modulators of androgen and estrogen receptor activity. Crit Rev Eukaryot Gene Expr 20:275-294.
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, et al. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927-937.
- Minutolo F, Macchia M, Katzenellenbogen BS, Katzenellenbogen JA (2011) Estrogen receptor beta ligands: recent advances and biomedical applications. Med Res Rev 31:364-372.
- Negro A, Brar BK, Gu Y, Peterson KL, Vale W (2006) ERB2 is required for G protein-coupled receptor signaling in the heart. ProcNatlAcadSci 103:155889-155893.
- Liu X, Gu X, Li Z, Li X, Li H, et al. (2006) Neuregulin-1/erbB-activation improves cardiac function and survival in models of ischemic, dilated, and viral cardiomyopathy. J Am Coll Cardiol 48:1438-1447.
- Rohrbach S, Niemann B, Silber RE, Holtz J (2005)Neuregulin receptors erbB2 and erbB4 in failing human myocardium -- depressed expression and attenuated activation. Basic Res Cardiol100:240-249.
- Rajagopalan V, Zucker IH, Jones JA, Carlson M, Ma YJ (2008) Cardiac ErbB-1/ErbB-2 mutant expression in young adult mice leads to cardiac dysfunction. Am J Physiol Heart Circ Physiol 295:543-554.
- Casademunt E, Carter BD, Benzel I, Frade JM, Dechant G, et al. (1999) The zinc finger protein NRIF interacts with the neurotrophin receptor p75(NTR) and participates in programmed cell death. EMBO J18:6050-6061.
- Tanigaki K, Honjo T (2007) Regulation of lymphocyte development by Notch signaling. Nat Immunol 8:451-456.
- Houghton AN (1994) Cancer antigens: immune recognition of self and altered self. J Exp Med 180:1-4.
- Houghton AN, Guevara-Patino JA (2004) Immune recognition of self in immunity against cancer. J Clin Invest 114:468-471.
- Tuohy VK, Lu Z, Sobel RA, Laursen RA, Lees MB (1989) Identification of an encephalitogenic determinant of myelin proteolipid protein for SJL mice. J Immunol 142:1523-1527.
- Bonavida B (1983) The SJL/J spontaneous reticulum cell sarcoma: new insights in the fields of neoantigens, host-tumor interactions, and regulation of tumor growth. Adv Cancer Res 38:1-22.
Supporting Information
Supplementary Figure Legends
Figure S-1: Comparable T cell responses between male and female SJL mice. (a) Proliferative response.
Male and female SJL mice were immunized with PLP 139-151 emulsified in CFA, and after 10 days, animals were euthanized to harvest the draining lymph nodes to prepare LNCs. Cells were stimulated with PLP 139-151 or a control peptide (NASE 101-120) for two days, and after pulsing with [H] thymidine for 16 hours, cells were harvested to measure the proliferative responses as cpm. Mean ± SEM values from three experiments involving one mouse each are shown. (b) Dextramer staining of T cells in vitro. LNCs stimulated as above were maintained in IL-2 medium, and the viable cells harvested on day 6 poststimulation were stained with PLP 139-151 or control (TMEV 70-86) dextramers for two hours followed by staining with anti-CD4 and 7-AAD. After washing, cells were acquired by flow cytometry, and the percentages of dextramer+ cells were determined in the live (7-AAD- ) CD4 T cell subset using FlowJo software. Flow cytometric density plots from one of the three experiments involving one to two mice each are shown. (c) Dextramer staining of braininfiltrates ex vivo. EAE was induced in male and female SJL mice by immunizing the animals with PLP 139-151 in CFA. The animals showing paralytic signs were euthanized and the brains were collected to harvest MNCs by percoll-gradient centrifugation method. Cells were stained with PLP 139-151 or control dextramers followed by anti-CD4 and 7-AAD, and the percentages of dextramer+ cells in the live subset (7-AAD-) were then determined by flow cytometry as described above. Representative flow cytometric density plots from three experiments are shown.
Figure 2: Crisscross analysis of the uniquely expressed proteins between males and females by IPA.
Protein samples derived from PLP 139-151-specific T cells of male and female mice were subjected to MS/MS analysis, and spectral counts were obtained for a list of proteins. The proteins that were uniquely expressed in males (10) and females (4) were used to obtain networks corresponding to ESR-1 and ESR-2 and AR, respectively.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences